Dr. Albitar’s molecular work is presented at the ASH meeting in oral and poster presentations.
Validation of Clinical Prognostic Models and Integration of Genetic Biomarkers of Drug Resistance in CLL Patients Treated with Ibrutinib
Program: Oral and Poster Abstracts | Monday, December 3, 2018: 11:45 AM
Type: Oral
Session: 642. CLL: Therapy, excluding Transplantation: Advances in CLL Using Novel Combination Therapy
Introduction:
We previously reported a prognostic scoring system in CLL using pre-treatment factors in patients treated with ibrutinib [Ahn et al, 2016 ASH Annual Meeting]. Here we present long-term follow-up results and validation of the prognostic models in a large independent cohort of patients. We also determine the incidence of resistance-conferring mutations in BTK and PLCG2 genes in different clinical risk groups.
Methods and Patients:
The discovery cohort comprised 84 CLL patients on a phase II study with either TP53 aberration (deletion 17p or TP53 mutation) or age ≥65 years (NCT01500733). The validation cohort comprised 607 patients pooled from four phase II and III studies for ibrutinib in treatment-naïve or relapsed/refractory CLL (NCT01105247; NCT01578707; NCT01722487; NCT01744691). All patients received single-agent ibrutinib 420mg once daily. We used Cox regression models to identify independent predictors of PFS, Kaplan-Meier method to estimate probabilities of PFS, log-rank test to compare PFS, and Cochran-Armitage trend test to compare the incidence of mutation among subgroups. We used R version 3.5.0 or SAS® version 9.3 for statistical analyses. For biomarker correlation, we tested cellular DNA or cell-free DNA collected from patients in the discovery cohort with the targeted sequencing of BTK and PLCG2 genes.
Result:
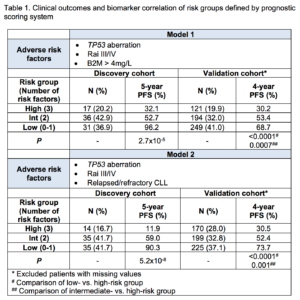
At a median follow-up of 5.2 years, 28 (33.3%) of 84 patients in the discovery cohort progressed or died. 52 (61.9%) patients had treatment-naïve CLL. Independent factors of PFS on univariate analysis were; TP53 aberration, prior treatment, and β-2 microglobulin (B2M) >4mg/L (P<0.05 for all tests). Unmutated IGHV and advanced Rai stage (III/IV) showed a trend toward inferior outcome without reaching statistical significance. Because higher levels of B2M were associated with relapsed/refractory CLL, we performed two multivariate Cox regression models to assess B2M and prior treatment status separately. Risk groups were determined by the presence of TP53 aberration, advanced Rai stage, and B2M >4mg/L for Model 1, and TP53 aberration, advanced Rai stage, and relapsed/refractory CLL for Model 2 (Table 1). The high-risk group had all three adverse risk factors; the intermediate-risk group had two risk factors; and the low-risk group, none or one. The median PFS of the high-risk group was 38.9 months for Model 1 and 38.4 months for Model 2, and was significantly shorter than those of intermediate and low-risk groups.
In the validation cohort, 254 (41.8%) of 607 patients progressed or died at a median follow-up of 4.2 years. 167 (27.5%) patients had treatment-naïve CLL. Both models showed statistically significant differences in PFS by risk groups (Table 1). For the high-risk group, 4-year PFS was 30.2% in Model 1 and 30.5% in Model 2, which were inferior to those of intermediate (53.4 and 52.4%) and low-risk groups (68.7 and 73.7%). Model 1 classified 20% of patients and Model 2 classified 28% of patients to the high-risk group.
BTK and PLCG2 mutations are common genetic drivers of ibrutinib resistance in CLL. To determine whether the incidence of these mutations correlates with prognostic risk groups, we performed targeted sequencing of BTK and PLCG2 of samples collected from patients in the discovery cohort. We used cell-free DNA for patients who received long-term ibrutinib (≥3 years) and had low circulating tumor burden, and cellular DNA, for samples collected within 3 years on ibrutinib or at progression. Of 84 patients, 69 (82.1%) were tested at least once, and 37 (44.0%) were tested at least twice. The frequency of testing was similar across the risk groups by two models (P>0.05). The cumulative incidences of mutations at 5 years in the low-, intermediate-, and high-risk groups were: 21.4%, 44.8% and 50%, respectively, by Model 1 (P=0.02); and 22.6%, 41.4% and 66.7%, respectively, by Model 2 (P=0.01).
Conclusion:
We developed and validated prognostic models to predict the risk of disease progression or death in CLL patients treated with ibrutinib. Risk groups classified by three commonly available pre-treatment factors showed statistically significant differences in PFS. The clinically-defined high-risk disease was linked to higher propensity to develop clonal evolution with BTK and/or PLCG2 mutations, which heralded ibrutinib resistance.
Inhye E. Ahn, MD1, Xin Tian2*, Maher Albitar, MD3*, Sarah E. M. Herman, PhD4*, Erika M. Cook4*, Susan Soto4*, Wanlong Ma5*, David Ipe6*, L. Claire Tsao6*, Mei Cheng6*, James P. Dean6 and Adrian Wiestner, MD7
1Hematology Branch, National Heart, Lung, and Blood Institute, National Institutes of Health, Rockville, MD
2Office of Biostatistics Research, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD
3Former: NeoGenomics, Valley Center, CA – Now: Genomic Testing Cooperative, Irvine, CA
4Hematology Branch, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD
5NeoGenomics Laboratories, Irvine, CA
6Pharmacyclics LLC, an AbbVie Company, Sunnyvale, CA
7NIH, NHLBI, Bethesda, MD