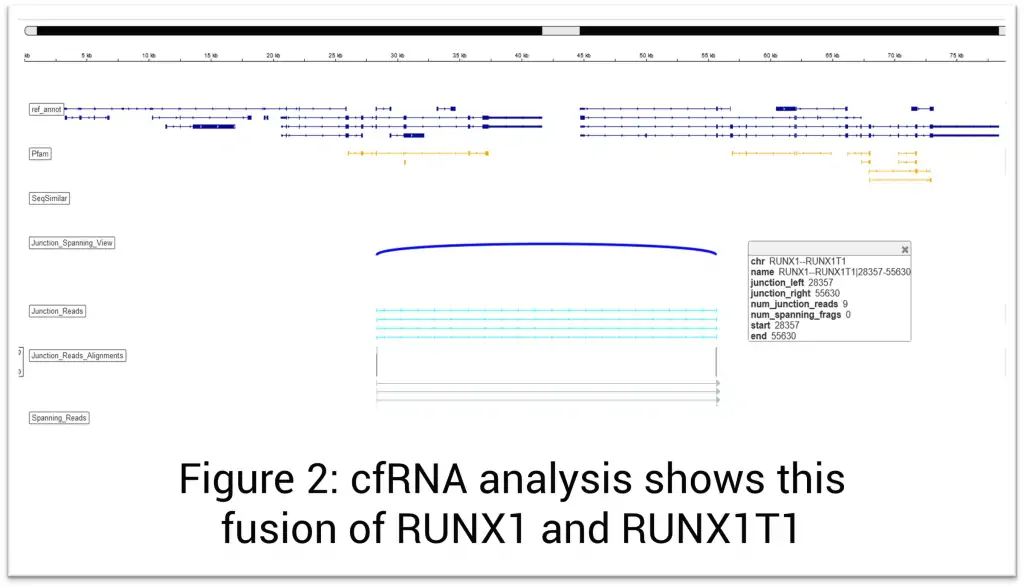
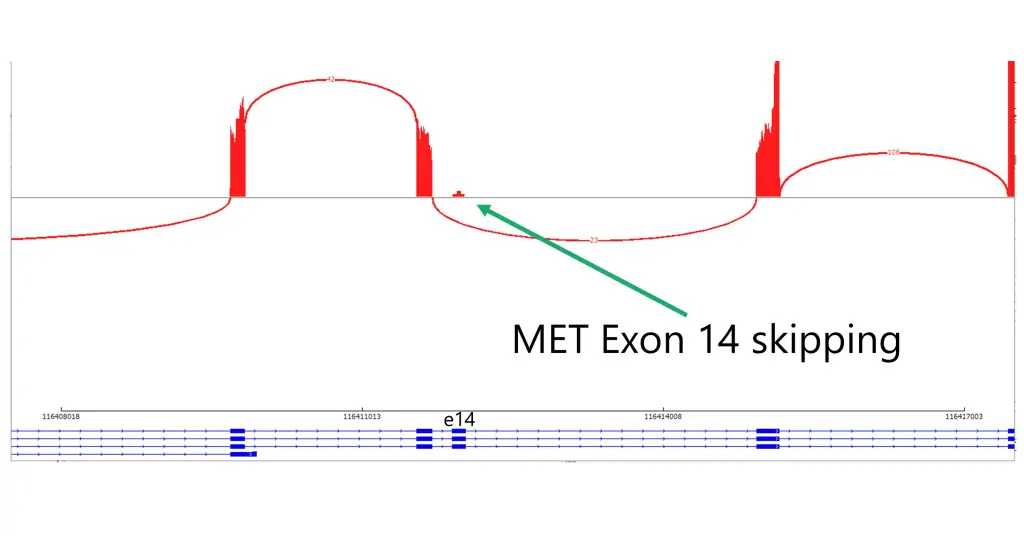
The Solid Tumor Profile Plus test combines the analysis of >400 DNA genes with targeted transcriptome sequencing (RNA) of more than 1600 genes to provide a comprehensive evaluation of cancer that includes detection of single-nucleotide variation, copy-number variation, gene expression levels, and fusions irrespective of their partner genes. In addition, the test is designed to detect microsatellite instability (MSI), tumor mutation burden (TMB), homologous recombination repair (HRR) mutations, and homologous recombination deficiency (HRD). Other notable features include evaluation for MET exon 14 skipping, EGFRvIII, AR-V7, TERT promoter mutations, DYPD gene polymorphism for prediction of toxicity to fluoropyrimidine therapy, and RNA levels of CTLA4, PD-L1, and PD-L2. The provided information helps in determining prognosis, designing a therapeutic approach, and predicting response to immunotherapies, targeted therapies, and precision medicines. It may also aid in biomarker discovery.
Targeted transcriptome sequencing can also detect:
- Gene expression levels that correlate to immunophenotype
- Gene amplifications
- Exon skipping
- Alternative splicing
- HLA class I genotyping
- Viral infections (EBV, HPV, TTV)
Epstein-Barr Virus (EBV): Important for diagnosis and classification of lymphoid neoplasms and some solid tumors.
Human Papillomavirus (HPV): Chronic infection with high-risk HPV subtypes is associated with increased risk of anogenital and oropharyngeal cancer. Detection of HPV mRNA suggests active (productive) infection.
Torque Teno Virus (TTV): This virus was first discovered in a patient with non-A-E hepatitis and is now regarded as a part of the human virome. In general, TTV does not cause pathology in immunocompetent individuals. This virus is considered as a marker of the degree of immune competence in patients with immunological impairment and inflammatory disorders. High TTV load is associated with increased risk of infection. In patients with organ transplant, low TTV load is associated with an increased risk of rejection.
T-cell and B-cell Clonality Detection: The detection of T- and B-cell clonality is important because it can help diagnose and monitor certain types of malignancies. When a malignant transformation occurs in a T- or B-cell, the cells can undergo uncontrolled clonal expansion, resulting in the accumulation of many cells with the identical T- or B-cell receptor.
MGMT Methylation: This assay is available as an add-on for brain cancer patients. MGMT methylation is predictive of response to radiotherapy and prognostic for glioblastoma.
Sensitivity is 3% for non-hotspot and 1% for hotspots. It is at 0.001 for cases with prior Hx.
Turn Around Time: 7-10 Days
GTC uses AI in every step of our analysis and it makes a difference in helping make a new discovery daily that improves patient care.
Once the data is offloaded from the sequencer, our AI:
- Assists with mutation analysis, identifying non-mutations and artifacts
- Compares various data sets to explore disease biology
- Provides support for clinical decision-making and classification of the disease
- It helps with matching patients to therapeutics and presents clinical trial options
- Aggregates data for report generation and simplifies the results so they are easily understood
Case Study: Cancer of Unknown Primary
Background
Cancer of unknown primary (CUP), when the site of tumor origin cannot be determined using the standard histological diagnostic tests, occurs in 3%–5% of cancer patients. Typically, patients with a CUP diagnosis are treated empirically and have inferior outcomes, with median overall survival less than one year. Gene expression profiling alone has been used to identify the tissue of origin but struggles with low neoplastic percentage in metastatic sites, where identification is often most needed. This case shows the value of AI paired with our comprehensive RNA profiling in determining cancers of CUP)/cell of origin.
Clinical History
- 61-year-old male with metastatic CUP in the femur
Molecular Profiling Findings
- Expression profile consistent with renal cell carcinoma
- Mutations in VHL, KDM5C, ARID2 (2 mutations), NIN, HGF, ARID1B, NTRK1, KMT2C, and EGFR (T790M) genes
- Chromosomal structural analysis shows 1q+, 3p-, 3q+, -4, 5q+ (distal), -9, 10p+, 10q-, -13, -14, and -21
- No evidence of microsatellite instability
- Tumor Mutation Burden High: 10 Mut/Mb
- Homologous recombination deficiency (HRD): Negative
Discussion
Using RNA and DNA sequencing and targeted transcriptome data coupled with machine learning, we were able to identify the kidney origin of the patient’s cancer along with detecting the VHL mutation, which has an FDA-approved targeted therapy (belzutifan). Our RNA profiling and artificial intelligence algorithms along with our experts interpterion of this information can provide highly useful tools for the pathologic diagnosis and classification of various cancers. The additional information that we provide, such as mutation profile and clinical information, can facilitate personalized treatment recommendations and minimize errors in pathologic diagnoses.
Download Case Study Brochure (PDF)
References
- Zhang H, Qureshi M, Wahid M, Charifa A, Ehsan A, IP A, De Dios I, Ma W, McCloskey J, Donato M, Siegel D, Gutierrez, Pecora A, Goy A, Albitar M. Differential diagnosis of hematologic and solid tumors using targeted Transcriptome and artificial intelligence (In press).
- Rassy E, Pavlidis N. Progress in refining the clinical management of cancer of unknown primary in the molecular era. Nat Rev Clin Oncol. 2020;17(9):541–554. doi: 10.1038/s41571-020-0359-1.
- Varadhachary GR, Raber MN. Cancer of unknown primary site. N Engl J Med. 2014;371(21):757–765. doi: 10.1056/NEJMc1411384.
- FFPE: 1 H&E slide and 6-8 unstained slides, 5-7 microns of tissue fixed with 10% NBF fixative. Please circle tumor for microdissection.
- Alternatively, the FFPE block can be sent for tumor circling and cutting at our laboratory.
Specimen Preparation and Shipping Guidelines
Use Solid Tumor Transport Kit
- Complete the requisition, making sure all sections are completed in their entirety including client information, patient information, specimen information, and test selection. Missing information may delay reporting of test results.
- Diagnosis/patient history is extremely important in rendering the correct interpretation of results and should also be filled out as completely as possible. A copy of a pathology report should be included.
- Ensure the specimen is labeled with patient name and number. A minimum of two patient identifiers is required for each specimen.
For FFPE samples:
- If sending FFPE blocks: Insert up to 6 blocks into the plastic block tray provided. Insert block tray into the foam insert in the transport box.
- If sending slides: Insert slides into the plastic slide holders provided. Insert the slide holders into the foam insert in the transport box. The container will hold up to 6 slide holders.
- Place folded test requisition(s), and/or manifest(s) if ordered electronically, into the transport box.
- Close the box and tuck the tabs into place. No tape is necessary.
Request Kits
Fill out the form below to request kits. Please refer to the Specimen Requirements page for more details.
*GTC will need to set you up in our system if this is your first order.
The Solid Tumor Profile Plus test combines the analysis of DNA with targeted transcriptome sequencing (RNA) to provide a comprehensive evaluation of cancer that includes detection of single nucleotide variation, copy number variation, gene expression levels and fusions irrespective of their partner genes. This includes testing of DNA
abnormalities in 434 genes and targeted transcriptome analysis of more than 1600 genes. In addition, the test is designed to detect microsatellite instability (MSI), tumor mutation burden (TMB), homologous recombination
repair (HRR) and homologous recombination deficiency (HRD). Other notable features include RNA levels of CTLA4, PD-L1, PD-L2, MET Exon 14 skipping, EGFRvIII, AR-V7 and DYPD gene polymorphism and prediction of toxicity to
fluoropyrimidine therapy. The provided information helps in determining prognosis, designing a therapeutic approach and predicting response to immunotherapies, targeted therapies, and precision medicines.
Targeted transcriptome sequencing can also detect:
• Gene expression levels that correlate to immunophenotype
• Gene amplifications
• Exon skipping
• Alternative splicing
• Biomarker discovery
We have recently added HPV testing to our Solid Tumor Profile Plus. HPV testing is useful for screening cervical and head and neck cancers. It can also be useful for monitoring patients and staging and screening of cervical lesions.
T-cell and B-cell clonality detection: The detection of T and B cell clonality is important because it can help diagnose and monitor certain types of malignancies. When a malignant transformation occurs in a T or B cell, the cells can undergo uncontrolled clonal expansion, resulting in the accumulation of a large number of identical cells with the same T or B cell receptor.
Turn Around Time: 7-10 Days
- FFPE: 1 H&E slide and 6-8 unstained slides, 5-7 microns of tissue fixed with 10% NBF fixative. Please circle tumor for microdissection. Alternatively, the FFPE block can be sent for tumor circling and cutting at our laboratory.
Specimen Preparation and Shipping Guidelines
Use Solid Tumor Transport Kit
- Complete Requisition, making sure all sections are completed in their entirety including client information, patient Information, specimen Information and test Selection. Missing information may delay reporting of test results.
- Diagnosis/patient history is extremely important in rendering the correct interpretation of results and should also be filled out as completely as possible. A copy of a Path report should be included.
- Ensure the specimen is labeled with patient name and number. A minimum of two patient identifiers is required for each specimen
For FFPE samples:
- If sending FFPE Block-Insert up to 6 blocks into plastic block tray provided. Insert block tray into foam insert in the transport box.
- If sending slides-Insert slides into plastic slide holders provided. Insert the slide holders into the foam insert in the transport box. The container will hold up to 6 maximum slide holders.
- Place folded test requisition(s) and/or manifest(s) if ordered electronically into the transport kit.
- Close box and tuck tabs into place. No tape necessary.
Genes validated and tested for Mutations in DNA testing for Solid Tumor
| 1-110 | 111-220 | 221-330 | 331-434 |
|---|---|---|---|
| ABCBA2:D1017 | EGLN1 | KDM6A | RAC1 |
| ABL1 | ELANE | KDR | RAD21 |
| ABL2 | EMSY | KEAP1 | RAD50 |
| ABRAXAS1 | EP300 | KEL | RAD51 |
| ACD | EPAS1 | KIF23 | RAD51B |
| ACVR1B | EPCAM | KIT | RAD51C |
| ADA | EPHA3 | KLF1 | RAD51D |
| ADGRA2 | EPHA5 | KLHL6 | RAD54L |
| AK2 | EPHA7 | KLLN | RAF1 |
| AKT1 | EPHB1 | KMT2A | RANBP2 |
| AKT2 | ERBB2 | KMT2B | RARA |
| AKT3 | ERBB3 | KMT2C | RB1 |
| ALK | ERBB4 | KMT2D | RBBP6 |
| AMER1 | ERCC4 | KRAS | RBM10 |
| ANKRD26 | ERG | LIG4 | RBM8A |
| APC | ERRFI1 | LMO1 | REEP5 |
| AR | ESR1 | LPIN2 | RET |
| ARAF | ETV6 | LRP1B | RHEB |
| ARFRP1 | EXO1 | LYN | RHOA |
| ARID1A | EZH2 | LYST | RICTOR |
| ARID1B | FANCA | LZTR1 | RIT1 |
| ARID2 | FANCB | MAGI2 | RNF168 |
| ASXL1 | FANCC | MAP2K1 | RNF43 |
| ATG2B | FANCD2 | MAP2K2 | ROS1 |
| ATM | FANCE | MAP2K4 | RPTOR |
| ATR | FANCF | MAP3K1 | RTEL1 |
| ATRX | FANCG | MAP3K14 | RUNX1 |
| AURKA | FANCI | MAPK1 | RUNX1T1 |
| AURKB | FANCL | MCL1 | SAMD9L |
| AURKC | FANCM | MDM2 | SBDS |
| AXIN1 | FAS | MDM4 | SBF2 |
| AXIN2 | FAT1 | MED12 | SDHA |
| AXL | FBXW7 | MEF2B | SDHB |
| B2M | FGF10 | MEFV | SDHC |
| BAP1 | FGF14 | MEN1 | SDHD |
| BARD1 | FGF19 | MET | SEC23B |
| BCL2 | FGF23 | MITF | SETBP1 |
| BCL2L1 | FGF3 | MLH1 | SETD2 |
| BCL2L2 | FGF4 | MPL | SF3B1 |
| BCL6 | FGF6 | MRE11 | SLIT2 |
| BCOR | FGFR1 | MSH2 | SLX4 |
| BCORL1 | FGFR2 | MSH6 | SMAD2 |
| BCR | FGFR3 | MTOR | SMAD3 |
| BIRC3 | FGFR4 | MUTYH | SMAD4 |
| BLM | FH | MVK | SMAD9 |
| BMPR1A | FLCN | MYC | SMARCA4 |
| BRAF | FLI1 | MYCL | SMARCB1 |
| BRCA1 | FLT1 | MYCN | SMC1A |
| BRCA2 | FLT3 | MYD88 | SMC3 |
| BRD4 | FLT4 | NBN | SMO |
| BRIP1 | FOXL2 | NF1 | SNCAIP |
| BTG1 | FOXP1 | NF2 | SOCS1 |
| BTK | FRS2 | NFE2L2 | SOX10 |
| CALR | FUBP1 | NFKBIA | SOX2 |
| CARD11 | G6PC3 | NHP2 | SOX9 |
| CBFB | GABRA6 | NKX2-1 | SPEN |
| CBL | GALNT12 | NLRP3 | SPOP |
| CBLB | GATA1 | NME1 | SPTA1 |
| CBLC | GATA2 | NOP10 | SRC |
| CCN6(WISP3) | GATA3 | NOTCH1 | SRSF2 |
| CCND1 | GATA4 | NOTCH2 | STAG2 |
| CCND2 | GATA6 | NOTCH3 | STAT3 |
| CCND3 | GEN1 | NPM1 | STAT4 |
| CCNE1 | GFI1 | NR0B1 | STAT6 |
| CD274 | GFI1B | NRAS | STK11 |
| CD79A | GID4 | NSD1 | SUFU |
| CD79B | GLI1 | NSD2(WHSC1) | SUZ12 |
| CDAN1 | GLI2 | NTRK1 | SYK |
| CDC73 | GNA11 | NTRK2 | TAF1 |
| CDH1 | GNA13 | NTRK3 | TAL1 |
| CDIN1(C15orf41) | GNAQ | NUP93 | TBX3 |
| CDK12 | GNAS | PAK3 | TCF3 |
| CDK4 | GREM1 | PALB2 | TCIRG1 |
| CDK6 | GRIN2A | PAX5 | TENT5C(FAM46C) |
| CDK8 | GRM3 | PBRM1 | TERC |
| CDKN1A | GSK3B | PDCD1LG2 | TERF1 |
| CDKN1B | GSKIP | PDGFRA | TERF2 |
| CDKN2A | H3-3A(H3F3A) | PDGFRB | TERF2IP |
| CDKN2B | H3C2 | PDK1 | TERT |
| CDKN2C | HAX1 | PHF6 | TET2 |
| CEBPA | HGF | PIK3C2B | TGFBR2 |
| CHD2 | HNF1A | PIK3CA | TNFAIP3 |
| CHD4 | HOXA11 | PIK3CB | TNFRSF14 |
| CHEK1 | HOXB13 | PIK3CG | TNFRSF1A |
| CHEK2 | HRAS | PIK3R1 | TOP1 |
| CIC | HSD3B1 | PIK3R2 | TOP2A |
| CREBBP | HSP90AA1 | PIM1 | TP53 |
| CRKL | ID3 | PLCG1 | TRAF3 |
| CRLF2 | IDH1 | PLCG2 | TSC1 |
| CSF1R | IDH2 | PMS1 | TSC2 |
| CSF3R | IGF1R | PMS2 | TSHR |
| CTC1 | IGF2 | POLD1 | U2AF1 |
| CTCF | IKBKE | POLE | U2AF2 |
| CTNNA1 | IKZF1 | POT1 | VEGFA |
| CTNNB1 | IKZF3 | PPM1D | VHL |
| CUL3 | IL2RG | PPP2R1A | WAS |
| CUX1 | IL7R | PRDM1 | WT1 |
| CXCR4 | INHBA | PREX2 | XPO1 |
| CYLD | INPP4B | PRKAR1A | XRCC2 |
| DAXX | IRF2 | PRKCI | XRCC3 |
| DDR2 | IRF4 | PRKDC | ZBTB2 |
| DDX11 | IRS2 | PRKN(PARK2) | ZNF217 |
| DDX41 | JAGN1 | PRSS1 | ZNF703 |
| DICER1 | JAK1 | PRSS8 | ZRSR2 |
| DKC1 | JAK2 | PSTPIP1 | |
| DNM2 | JAK3 | PTCH1 | |
| DNMT3A | JUN | PTEN | |
| DOT1L | KAT6A | PTPN11 | |
| EED | KDM5A | QKI | |
| EGFR | KDM5C | RAB27A |
(* Microsatellite markers BAT25, BAT26, D2S123, D5S346, and D17S250 are included.)
More than 1600 genes in total validated and tested for mutations in cfRNA testing
IMGT repertoire comprises of genes in : IgK, IgL, IgH, T-Receptor A, T-Receptor B, T-Receptor D, and T-Receptor G genes
| 1-317 | 318-634 | 635-951 | 952-1268 | 1269-1584 |
|---|---|---|---|---|
| ABCB1 | CREB3L1 | HDAC5 | NAPSA | SDHC |
| ABCC2 | CREB3L2 | HDAC6 | NAV3 | SDHD |
| ABCC3 | CREBBP | HDAC7 | NBN | SEC31A |
| ABCG2 | CRKL | HECW1 | NBR1 | SEPTIN2 (SEPT2) |
| ABI1 | CRLF2 | HEPH | NCAM1 (CD56) | SEPTIN5 (SEPT5) |
| ABL1 | CRTC1 | HERPUD1 | NCKIPSD | SEPTIN6 (SEPT6 ) |
| ABL2 | CRTC3 | HES1 | NCOA1 | SEPTIN9 (SEPT9) |
| ABLIM1 | CSF1 | HES5 | NCOA2 | SERP2 |
| ABRAXAS1 (FAM175A) | CSF1R | HEY1 | NCOA3 | SERPINE1 |
| ACACA | CSF3 | HGF | NCOA4 | SERPINF1 |
| ACE | CSF3R | HHEX | NCOR2 | SET |
| ACER1 | CSNK1G2 | HIF1A | NCR1 | SETBP1 |
| ACKR3 | CSNK2A1 | HIP1 | NCSTN | SETD2 |
| ACP3 | CTCF | HIPK1 | NDC80 | SETD7 |
| ACSBG1 | CTDSP2 | HIPK2 | NDE1 | SF3A1 |
| ACSL3 | CTLA4 | HLF | NDRG1 | SF3B1 |
| ACSL6 | CTNNA1 | HMGA1 | NDUFAF1 | SFPQ |
| ACVR1 | CTNNB1 | HMGA2 | NECTIN4 (PVRL4) | SFRP2 |
| ACVR1B | CTNND2 | HMGB1 | NEDD4 | SFRP4 |
| ACVR1C | CTRB1 | HNF1A | NEURL1 | SGK1 |
| ACVR2A | CTRB2 | HNRNPA2B1 | NEUROD1 | SGPP2 |
| ACVRL1 | CTSA | HOOK3 | NF1 | SH2D5 |
| ADD3 | CUX1 | HOXA10 | NF2 | SH3BP1 |
| ADGRA2 | CXCL8 | HOXA11 | NFATC1 | SH3D19 |
| ADGRG7 | CXCR4 | HOXA13 | NFATC2 | SH3GL1 |
| ADM | CXXC4 | HOXA3 | NFE2 | SH3GL2 |
| AFDN (MLLT4) | CYFIP2 | HOXA5 | NFE2L2 | SHC1 |
| AFF1 | CYLD | HOXA9 | NFIB | SHC2 |
| AFF3 | CYP1B1 | HOXC11 | NFKB1 | SHTN1 |
| AFF4 | CYP2C19 | HOXC13 | NFKB2 | SIK3 |
| AFP | DAB2IP | HOXD11 | NFKBIA | SIN3A |
| AGR3 | DACH1 | HOXD13 | NFYC | SIRT1 |
| AHCYL1 | DACH2 | HOXD9 | NGF | SKP2 |
| AHI1 | DAXX | HRAS | NGFR | SLAMF7 |
| AHR | DCLK2 | HSP90AA1 | NIN | SLC1A2 |
| AIP | DCN | HSP90AB1 | NIPBL | SLC22A1 |
| AK2 | DDB2 | HSPA1A | NKX2-1 | SLC22A2 |
| AK5 | DDIT3 | HSPA1B | NKX2-5 | SLC34A2 |
| AKAP12 | DDR2 | HSPA2 | NKX3-1 | SLC45A3 |
| AKAP6 | DDX10 | HSPA4 | NOD1 | SLC47A1 |
| AKAP9 | DDX20 | HSPA5 | NODAL | SLC47A2 |
| AKR1C3 | DDX39B | HTRA1 | NONO | SLC66A3 (PQLC3) |
| AKT1 | DDX3X | HUWE1 | NOS3 | SLC7A5 |
| AKT2 | DDX41 | IBSP | NOTCH1 | SLCO1B1 |
| AKT3 | DDX5 | ICAM1 | NOTCH2 | SLCO1B3 |
| ALDH1A1 | DDX6 | ICOS | NOTCH3 | SLX4 |
| ALDH2 | DEK | ID1 | NOTCH4 | SMAD2 |
| ALDOC | DGKB | ID3 | NPM1 | SMAD3 |
| ALK | DGKI | ID4 | NPM2 | SMAD4 |
| AMER1 | DGKZ | IDH1 | NR3C1 | SMAD5 |
| AMH | DICER1 | IDH2 | NR4A3 | SMAD6 |
| ANGPT1 | DIRAS3 | IDO1 | NR5A1 | SMAP1 |
| ANKRD26 | DIS3L2 | IFNA1 | NR6A1 | SMARCA1 |
| ANKRD28 | DKK1 | IFNA2 | NRAS | SMARCA4 |
| ANLN | DKK2 | IFNG | NRG1 | SMARCA5 |
| ANPEP (CD13) | DKK4 | IFRD1 | NSD1 | SMARCB1 |
| APC | DLEC1 | IGF1 | NSD2 (WHSC1) | SMC1A |
| APH1A | DLL1 | IGF1R | NSD3 (WHSC1L1) | SMC3 |
| APLP2 | DLL3 | IGFBP2 | NT5C2 | SMO |
| APOD | DLL4 | IGFBP3 | NTF3 | SNAPC3 |
| AR | DMRT1 | IGLL5 | NTF4 | SNCG |
| ARAF | DMRTA2 | IKBKB | NTHL1 | SNW1 |
| ARFRP1 | DNAJB1 | IKBKE | NTRK1 | SNX29 |
| ARG1 | DNM1 | IKZF1 | NTRK2 | SNX9 |
| ARHGAP20 | DNM2 | IKZF2 | NTRK3 | SOCS1 |
| ARHGAP26 | DNM3 | IKZF3 | NUMA1 | SOCS2 |
| ARHGEF12 | DNMT1 | IL10 | NUP107 | SOCS3 |
| ARHGEF7 | DNMT3A | IL12RB2 | NUP214 | SOD2 |
| ARID1A | DNTT (TdT) | IL13 | NUP93 | SORBS2 |
| ARID2 | DOCK1 | IL13RA2 | NUP98 | SORT1 |
| ARIH2 | DOT1L | IL15 | NUTM1 | SOS1 |
| ARNT | DPM1 | IL15RA | NUTM2A | SOX10 |
| ARRDC4 | DPP4 | IL17A | NUTM2B | SOX11 |
| ASCL1 | DPYD | IL17B | OCLN | SOX2 |
| ASMTL | DST | IL1B | OFD1 | SP1 |
| ASPH | DTX1 | IL1R1 | OGA (MGEA5) | SP3 |
| ASPSCR1 | DTX4 | IL1RAP | OLIG1 | SPECC1 |
| ASTN2 | DUSP2 | IL2 | OLIG2 | SPEN |
| ASXL1 | DUSP22 | IL21R | OLR1 | SPN |
| ATF1 | DUSP26 | IL2RA (CD25) | OMD | SPOP |
| ATF3 | DUSP9 | IL3 | OPN1LW | SPP1 |
| ATG13 | DUX4 | IL33 | P2RY8 | SPRY2 |
| ATG5 | E2F1 | IL36G | PAFAH1B2 | SPRY4 |
| ATIC | EBF1 | IL3RA (CD123) | PAG1 | SPTAN1 |
| ATL1 | ECT2L | IL6 | PAK1 | SPTBN1 |
| ATM | EDIL3 | IL7R | PAK3 | SQSTM1 |
| ATP1B4 | EDNRB | INHBA | PAK5 (PAK7) | SRC |
| ATP6V1G2-DDX39B | EED | INPP4A | PAK6 | SRF |
| ATP8A2 | EEFSEC | INPP4B | PALB2 | SRGAP3 |
| ATR | EGF | INPP5A | PAPPA | SRRM3 |
| ATRNL1 | EGFR | INPP5D | PASK | SRSF2 |
| ATRX | EGR1 | IQCG | PATZ1 | SRSF3 |
| AURKA | EGR2 | IRAG2 (LRMP) | PAX3 | SS18 |
| AURKB | EGR3 | IRF1 | PAX5 | SS18L1 |
| AUTS2 | EGR4 | IRF2BP2 | PAX7 | SSBP2 |
| AXIN1 | EIF4A2 | IRF4 (MUM1) | PAX8 | SSX1 |
| AXIN2 | EIF4E | IRF8 | PBRM1 | SSX2 |
| AXL | ELANE | IRS1 | PBX1 | SSX2B |
| B2M | ELF4 | IRS2 | PBX3 | SSX4 |
| B3GAT1 (CD57) | ELK4 | IRS4 | PC | SSX4B |
| BACH1 | ELL | ITGA2B (CD41) | PCA3 | ST3GAL4 |
| BACH2 | ELN | ITGA5 | PCBP1 | ST6GAL1 |
| BAG4 | ELOVL2 | ITGA7 | PCLO | STAG2 |
| BAIAP2L1 | ELP2 | ITGA8 | PCM1 | STAT1 |
| BAP1 | EML1 | ITGAE (CD103) | PCNA | STAT3 |
| BARD1 | EML4 | ITGAM (CD11B) | PCSK7 | STAT4 |
| BAX | EMSY | ITGAV | PDCD1 (PD-1/CD279) | STAT5A |
| BAZ2A | ENG | ITGAX (CD11C) | PDCD11 | STAT5B |
| BCAS3 | ENPP2 | ITGB3 (CD61) | PDCD1LG2 (PD-L2) | STAT6 |
| BCAS4 | ENTPD1 | ITGB4 | PDCD6-AHRR | STIL |
| BCL10 | EP300 | ITK | PDE4DIP | STK11 |
| BCL11A | EP400 | ITPKA | PDGFA | STRN |
| BCL11B | EPC1 | JAG2 | PDGFB | STX5 |
| BCL2 | EPCAM | JAK1 | PDGFD | STYK1 |
| BCL2A1 | EPHA10 | JAK2 | PDGFRA | SUFU |
| BCL2L1 | EPHA2 | JAK3 | PDGFRB | SUGP2 |
| BCL2L2 | EPHA3 | JARID2 | PDK1 | SULF1 |
| BCL3 | EPHA5 | JAZF1 | PEG3 | SUV39H2 |
| BCL6 | EPHA7 | JUN | PER1 | SUZ12 |
| BCL7A | EPHB1 | KALRN | PFDN5 | SYK |
| BCL9 | EPHB6 | KAT6A | PHB | SYP |
| BCOR | EPO | KAT6B | PHF1 | TACC1 |
| BCORL1 | EPOR | KCNB1 | PHF19 | TACC2 |
| BCR | EPS15 | KDM1A | PHF23 | TACC3 |
| BDNF | ERBB2 | KDM2B | PHF6 | TACSTD2 (TROP2) |
| BHLHE22 | ERBB3 | KDM4C | PHOX2B | TAF1 |
| BICC1 | ERBB4 | KDM5A | PI4KA | TAF15 |
| BIN1 | ERC1 | KDM5C | PICALM | TAFA2 (FAM19A2) |
| BIRC3 | ERCC1 | KDM6A | PIGA | TAFA5 (FAM19A5) |
| BIRC6 | ERCC2 | KDR | PIK3CA | TAL1 |
| BLM | ERCC3 | KDSR | PIK3CB | TAL2 |
| BMP4 | ERCC4 | KEAP1 | PIK3CD | TAOK1 |
| BMPR1A | ERCC5 | KIAA0232 | PIK3CG | TBL1XR1 |
| BRAF | ERCC6 | KIAA1549 | PIK3R1 | TBX15 |
| BRCA1 | ERCC6L2 | KIF5B | PIK3R2 | TBX21 |
| BRCA2 | ERG | KIR3DL1 | PIM1 | TCEA1 |
| BRD1 | ERLIN2 | KIT (CD117) | PIMREG (FAM64A) | TCF12 |
| BRD3 | ESR1 | KLF2 | PITX2 | TCF3 |
| BRD4 | ETNK1 | KLF4 | PIWIL1 | TCF7 |
| BRIP1 | ETS1 | KLHL6 | PKM | TCF7L2 |
| BRSK1 | ETS2 | KLK2 | PLA2G2A | TCL1A |
| BRWD3 | ETV1 | KLK3 (PSA) | PLA2G5 | TCL1B |
| BTBD18 | ETV4 | KLK7 | PLAG1 | TCL6 |
| BTG1 | ETV5 | KLRC1 | PLAT | TCTA |
| BTG2 | ETV6 | KMT2A | PLAU | TEAD1 |
| BTK | EVI2A | KMT2B | PLCB1 | TEAD2 |
| BTLA | EVI2B | KMT2C | PLCB4 | TEAD3 |
| BUB1B | EWSR1 | KMT2D | PLCG1 | TEAD4 |
| C10orf55 | EXO1 | KNL1 (CASC5) | PLCG2 | TEC |
| C11orf1 | EXOSC6 | KPNB1 | PLEKHM2 | TENM1 |
| C11orf54 | EXT1 | KRAS | PLPP3 | TENT5C (FAM46C) |
| C2CD2L | EXT2 | KRT1 | PML | TERC |
| CACNA1F | EYA1 | KRT10 | PMS1 | TERF1 |
| CACNA1G | EYA2 | KRT16 | PMS2 | TERF2 |
| CACNA2D3 | EZH2 | KRT17 | POFUT1 | TERT |
| CAD | EZR | KRT19 | POLD1 | TET1 |
| CALR | F3 (CD142) | KRT2 | POLD4 | TET2 |
| CAMK2A | FAF1 | KRT5 | POLE | TFDP1 |
| CAMK2B | FANCA | KRT6A | POLR2H | TFE3 |
| CAMK2G | FANCB | KRT6B | POM121 | TFEB |
| CAMTA1 | FANCC | KRT8 | POMGNT1 | TFG |
| CANT1 | FANCD2 | KSR1 | POSTN | TFPT |
| CAPRIN1 | FANCE | KTN1 | POT1 | TFRC (CD71) |
| CAPZB | FANCF | LAG3 | POU2AF1 (BOB1) | TG |
| CARD11 | FANCG | LAMA1 | POU2F3 | TGFB2 |
| CARM1 | FANCI | LAMA5 | POU5F1 | TGFB3 |
| CARMIL2 (RLTPR) | FANCL | LAMP1 (CD107A) | PPARG | TGFBI |
| CARS1 (CARS) | FANCM | LAMP2 | PPARGC1A | TGFBR2 |
| CASP3 | FAS | LASP1 | PPFIA2 | TGFBR3 |
| CASP7 | FASLG | LCK | PPFIBP1 | THADA |
| CASP8 | FBN2 | LCP1 | PPM1D | THBS1 |
| CAV1 | FBXO11 | LDHA | PPP1CB | THPO |
| CBFA2T3 | FBXO31 | LDHB | PPP1R13B | THRA |
| CBFB | FBXW7 | LDHC | PPP1R13L | THRAP3 |
| CBL | FCER2 (CD23) | LEF1 | PPP2CB | TIAM1 |
| CBLB | FCGBP | LEFTY2 | PPP2R1A | TIRAP |
| CBLC | FCGR1A (CD64) | LFNG | PPP2R1B | TLL2 |
| CCAR2 | FCGR2B | LGALS3 | PPP2R2B | TLR4 |
| CCDC170 | FCGR3A (CD16) | LGR5 | PPP3CA | TLX1 |
| CCDC28A | FCRL4 | LHFPL3 | PPP3CB | TLX3 |
| CCDC6 | FEN1 | LHFPL6 (LHFP) | PPP3CC | TMEM127 |
| CCDC88C | FEV | LHX2 | PPP3R1 | TMEM230 |
| CCK | FGF1 | LHX4 | PPP3R2 | TMEM30A |
| CCL2 | FGF10 | LIFR (CD118) | PPP4C | TMPRSS2 |
| CCNA2 | FGF13 | LILRA4 | PRAME | TNC |
| CCNB1IP1 | FGF14 | LINGO2 | PRCC | TNF |
| CCNB3 | FGF19 | LMBRD1 | PRDM1 | TNFAIP3 |
| CCND1 | FGF2 | LMO1 | PRDM16 | TNFRSF10B |
| CCND2 | FGF23 | LMO2 | PRDM7 | TNFRSF10D |
| CCND3 | FGF3 | LMO7 | PRF1 | TNFRSF11A |
| CCNE1 | FGF4 | LNP1 | PRG2 | TNFRSF13C (BAFFR) |
| CCNG1 | FGF6 | LOX | PRICKLE1 | TNFRSF14 |
| CCR4 | FGF8 | LPAR1 | PRKACA | TNFRSF17 (BCMA) |
| CCT6B | FGF9 | LPP | PRKACG | TNFRSF4 |
| CD14 | FGFR1 | LPXN | PRKAR1A | TNFRSF6B |
| CD157 (BST1) | FGFR1OP (CEP43) | LRIG3 | PRKCA | TNFRSF8 (CD30) |
| CD19 | FGFR1OP2 | LRP1B | PRKCB | TNFRSF9 (CD137) |
| CD1A | FGFR2 | LRP5 | PRKCD | TOP1 |
| CD2 | FGFR3 | LRPPRC | PRKCG | TOP2A |
| CD200 | FGFR4 | LRRC37B | PRKDC | TOP2B |
| CD22 | FH | LRRC59 | PRKG2 | TP53 |
| CD24 | FHIT | LRRC7 | PRMT1 | TP53BP1 |
| CD274 (PD-L1) | FHL2 | LRRK2 | PRMT8 | TP63 |
| CD276 (B7-H3) | FIP1L1 | LTBP1 | PROM1 | TP73 |
| CD28 | FLCN | LUC7L2 | PRPF40B | TPD52L2 |
| CD33 | FLI1 | LY75 | PRPF8 | TPM3 |
| CD34 | FLNA | LYL1 | PRRX1 | TPM4 |
| CD36 | FLNC | LYN | PRRX2 | TPO |
| CD37 | FLT1 | MACROD1 | PRSS3 | TPR |
| CD38 | FLT3 | MAD2L1 | PRSS8 | TRAF2 |
| CD3D | FLT3LG | MADD | PSD3 | TRAF3 |
| CD3E | FLT4 | MAF | PSEN1 | TRAF5 |
| CD3G | FLYWCH1 | MAFB | PSIP1 | TRHDE |
| CD3Z (CD247) | FNBP1 | MAGEA4 | PSMD2 | TRIM24 |
| CD4 | FOLH1 (PSMA) | MAGED1 | PTBP1 | TRIM27 |
| CD40 | FOLR1 | MAGEE1 | PTCH1 | TRIM33 |
| CD40LG | FOS | MAL | PTCRA | TRIP11 |
| CD44 | FOSB | MALT1 | PTEN | TRPS1 |
| CD47 | FOSL1 | MAML1 | PTGS2 | TSC1 |
| CD5 | FOXL2 | MAML2 | PTK2 | TSC2 |
| CD52 | FOXO1 | MAP2 | PTK2B | TSHR |
| CD58 | FOXO3 | MAP2K1 | PTK7 | TTF1 |
| CD59 | FOXO4 | MAP2K2 | PTPA (PPP2R4) | TTK |
| CD62L (SELL) | FOXP1 | MAP2K3 | PTPN11 | TTL |
| CD68 | FOXP3 | MAP2K4 | PTPN2 | TTYH1 |
| CD7 | FRK | MAP2K5 | PTPN6 | TUSC3 |
| CD70 | FRMPD4 | MAP2K6 | PTPRA | TYK2 |
| CD74 | FRS2 | MAP2K7 | PTPRC (CD45) | TYMS |
| CD79A | FRYL | MAP3K1 | PTPRD | U2AF1 |
| CD79B | FSTL3 | MAP3K14 | PTPRK | U2AF2 |
| CD81 | FUS | MAP3K6 | PTPRO | UBA1 |
| CD8A | FUT1 | MAP3K7 | PTPRR | UBE2B |
| CD8B | FUT4 (CD15) | MAPK1 | PTTG1 | UBE2C |
| CD9 | FUT7 | MAPK3 | RABEP1 | UBTF |
| CDA | FZD10 | MAPK8 | RAC1 | UFC1 |
| CDC14A | FZD2 | MAPK8IP2 | RAC2 | UFM1 |
| CDC14B | FZD3 | MAPK9 | RAC3 | UPK3A |
| CDC25A | FZD6 | MAPRE1 | RAD21 | USP16 |
| CDC25C | FZD7 | MATK | RAD50 | USP42 |
| CDC42 | FZD8 | MAX | RAD51 | USP5 |
| CDC73 | GAB1 | MB21D2 | RAD51B | USP6 |
| CDH1 | GABARAP | MBNL1 | RAD51C | USP7 |
| CDH11 | GABRG2 | MBTD1 | RAD51D | UTP4 |
| CDK1 | GADD45B | MCAM | RAD52 | VCAM1 |
| CDK12 | GALNT12 | MCL1 | RAF1 | VEGFA |
| CDK2 | GANAB | MCM3AP | RALGDS | VEGFC |
| CDK4 | GAS1 | MDC1 | RANBP17 | VEGFD (FIGF) |
| CDK5RAP2 | GAS7 | MDH1 | RANBP2 | VGLL3 |
| CDK6 | GATA1 | MDM2 | RAP1GDS1 | VHL |
| CDK7 | GATA2 | MDM4 | RARA | VPS45 |
| CDK8 | GATA3 | MDS2 | RASAL1 | VSIR (C10orf54) |
| CDK9 | GATA6 | MEAF6 | RASGEF1A | VTCN1 |
| CDKL5 | GBP1 | MECOM | RASGRF1 | VTI1A |
| CDKN1A | GBP2 | MED12 | RASGRF2 | WASF2 |
| CDKN1B | GDF6 | MEF2B | RASGRP1 | WDCP (C2orf44) |
| CDKN1C | GFAP | MEF2C | RB1 | WDFY3 |
| CDKN2A | GHR | MEF2D | RBM15 | WDR1 |
| CDKN2B | GID4 | MEIS1 | RBM6 | WDR18 |
| CDKN2C | GIT2 | MELK | RCHY1 | WDR70 |
| CDKN2D | GLI1 | MEN1 | RCOR1 | WDR90 |
| CDX1 | GLI3 | MET | RCSD1 | WEE1 |
| CDX2 | GMPS | METTL18 | RECQL4 | WIF1 |
| CEACAM5 (CEA) | GNA11 | METTL7B | REEP3 | WNT10A |
| CEACAM8 | GNA12 | MFNG | REG3A | WNT10B |
| CEBPA | GNA13 | MGMT | RELA | WNT11 |
| CEBPB | GNAI1 | MIB1 | RELN | WNT16 |
| CEBPD | GNAQ | MIPOL1 | RERG | WNT2B |
| CEBPE | GNAS | MITF | RET | WNT3 |
| CENPF | GNB1 | MKI67 | RGS7 | WNT4 |
| CENPU | GNG4 | MLANA | RHBDF2 | WNT5B |
| CEP170B | GOLGA5 | MLF1 | RHOA | WNT6 |
| CEP57 | GOPC | MLH1 | RHOD | WNT7B |
| CEP85L | GOSR1 | MLLT1 | RHOH | WNT8B |
| CHCHD7 | GOT1 | MLLT10 | RICTOR | WRN |
| CHD2 | GPC3 | MLLT11 | RMI2 | WSB1 |
| CHD6 | GPHN | MLLT3 | RNF213 | WT1 |
| CHEK1 | GPR34 | MLLT6 | RNF217-AS1 | WWOX |
| CHEK2 | GPRC5D | MME (CD10) | RNF43 | WWTR1 |
| CHIC2 | GRB10 | MMP7 | ROBO1 | XBP1 |
| CHL1 | GRB2 | MMP9 | ROBO2 | XIAP |
| CHMP2B | GREM1 | MN1 | ROR1 | XKR3 |
| CHN1 | GRHPR | MNAT1 | ROR2 | XPA |
| CHST11 | GRID1 | MNX1 | ROS1 | XPC |
| CHUK | GRIN2A | MPL | RPA3 | XPO1 |
| CIC | GRIN2B | MPO | RPL21 | XRCC2 |
| CIITA | GRM1 | MRE11 (MRE11A) | RPL22 | XRCC3 |
| CILK1 | GRM3 | MRTFA (MKL1) | RPN1 | XRCC6 |
| CIP2A (KIAA1524) | GSK3B | MRTFB (MKL2) | RPN2 | YAP1 |
| CIT | GSN | MS4A1 (CD20) | RPS21 | YPEL5 |
| CKB | GTF2I | MSC | RPS6KA1 | YTHDF2 |
| CKS1B | GTSE1 | MSH2 | RPS6KA2 | YWHAE |
| CLDN18 | GUCY2C | MSH3 | RPS6KA3 | YY1AP1 |
| CLEC12A | GYPA (CD235A) | MSH5 | RPTOR | ZAP70 |
| CLEC4C | GZMA | MSH6 | RREB1 | ZBTB16 |
| CLP1 | GZMB | MSI2 | RRM1 | ZC3H7A |
| CLTA | H1-2 (HIST1H1C) | MSN | RRM2B | ZC3H7B |
| CLTC | H1-3 (HIST1H1D) | MTAP | RTEL1 | ZFP64 |
| CLTCL1 | H1-4 (HIST1H1E) | MTCP1 | RTEL1-TNFRSF6B | ZFPM2 |
| CMKLR1 | H2AC11 | MTOR | RTL8B (FAM127C) | ZFTA (C11orf95) |
| CNBP | H2AC16 | MTUS2 | RTN3 | ZFYVE19 |
| CNOT2 | H2AC17 | MUC1 (CA15-3) | RUNX1 | ZIC2 |
| CNTN1 | H2AC6 | MUC16 (CA-125) | RUNX1T1 | ZMIZ1 |
| CNTRL | H2AX | MUTYH | RUNX2 | ZMYM2 |
| COG5 | H2BC11 | MYB | RYR3 | ZMYM3 |
| COL11A1 | H2BC12 | MYBL1 | S1PR2 | ZMYND11 |
| COL1A1 | H2BC17 | MYC | SAMD9 | ZNF207 |
| COL1A2 | H2BC4 | MYCL | SAMD9L | ZNF217 |
| COL3A1 | H2BC5 | MYCN | SARNP | ZNF24 |
| COL6A3 | H3-3A (H3F3A) | MYD88 | SATB2 | ZNF331 |
| COL9A3 | H3C2 | MYH11 | SBDS | ZNF384 |
| COMMD1 | H4C9 | MYH9 | SCGB2A2 | ZNF444 |
| COX6C | HAS2 | MYO18A | SCN8A | ZNF521 |
| CPNE1 | HAVCR2 | MYO1F | SDC1 (CD138) | ZNF585B |
| CPS1 | HDAC1 | NAB2 | SDC4 | ZNF687 |
| CPSF6 | HDAC2 | NACA | SDHA | ZNF703 |
| CRADD | HDAC3 | NAMPT | SDHAF2 | ZRSR2 |
| CREB1 | HDAC4 | NAPA | SDHB |
Request Kits
Fill out the form below to request kits. Please refer to the Specimen Requirements page for more details.
*GTC will need to set you up in our system if this is your first order.
GTC uses AI in every step of our analysis and it makes a difference in helping make a new discovery daily that improve patient care.
Once the data is offloaded from the sequencer, our AI:
- Assists with mutation analysis, identifying non-mutations and artifacts
- Compares various data sets to explore disease biology
- Provides support for clinical decision making and classification of the disease
- It helps with matching patients to therapeutics and presents clinical trial options
- Aggregates data for report generation and simplifies the results so they are easily understood
Case Study: Cancer of Unknown Primary
Background
Cancer of Unknown Primary (CUP) occurs in 3–5% of patients when the site of tumor origin cannot be determined using the standard histological diagnostic tests. Typically, a CUP diagnosis is treated empirically and has inferior outcomes, with median overall survival less than one year. Gene expression profiling alone has been used to identify the tissue of origin but struggles with low neoplastic percentage in metastatic sites which is where identification is often most needed. This case shows the value of AI paired with our comprehensive RNA profiling in determining Cancers of Unknown Primary (CUP)/Cell of Origin.
Clinical History
- 61-year-old male
- With metastatic cancer of unknown primary to the femur
Molecular Profiling Findings
- Expression profile consistent with renal cell carcinoma
- Mutations in VHL, KDM5C, ARID2 (2 mutations), NIN, HGF, ARID1B, NTRK1, KMT2C, EGFR(T790M) genes
- Chromosomal structural analysis shows 1q+, 3p-, 3q+, -4, 5q+ (distal), -9, 10p+, 10q-, -13, -14, and -21
- No evidence of microsatellite instability
- Tumor Mutation Burden High: 10 Mut/Mb
- Homologous recombination deficiency (HRD): Negative
Discussion
Using RNA and DNA sequencing and targeted transcriptome data coupled with machine learning; we were able to identify the kidney origin of the patient’s cancer along with detecting the VHL mutation which has an FDA approved targeted therapy (Belzutifan). Our RNA profiling and artificial intelligence algorithms along with our experts interpterion of this information can provide highly useful tools for the pathologic diagnosis and classification of various cancers. The additional information that we provide such as mutation profile and clinical information can provide personalized treatment recommendations and minimize errors in pathologic diagnoses.
Download Case Study Brochure (PDF)
References
- Zhang H, Qureshi M, Wahid M, Charifa A, Ehsan A, IP A, De Dios I, Ma W, McCloskey J, Donato M, Siegel D, Gutierrez, Pecora A, Goy A, Albitar M Differential Diagnosis of Hematologic and Solid tumors Using Targeted Transcriptome and Artificial Intelligence (In press)
- Rassy, E. & Pavlidis, N. Progress in refining the clinical management of cancer of unknown primary in the molecular era. Nat. Rev. Clin. Oncol. 17, 541–554 (2020).
- Varadhachary, G. R. & Raber, M. N. Cancer of unknown primary site. N. Engl. J. Med. 371, 757–765 (2014).
How to complete the Genomic Testing Cooperative requisition form.
Download our
Test Requisition
Keep in mind that we do not accept blood samples directly from individuals. Talk with your M.D. to fill out the form for you.
