GTC-Hematology Profile Plus combines expression and fusion with mutation analysis in DNA and RNA.

The test covers 302 DNA genes and more than 1600 RNA genes.
This is a comprehensive evaluation for all hematologic neoplasms.
However, it is especially recommended for:
Acute Myeloid Leukemia (AML): Translocations in AML are very important for diagnosis, prognosis and therapy selection. This comprehensive test can provide a complete evaluation of fusion mRNA and mutations. It also helps in determining a diagnosis in acute leukemia with ambiguous phenotype.
Clonal Hematopoiesis of Indeterminate Potential (CHIP): Distinguish CHIP from clinically active and relevant hematologic neoplasm based on an internally developed algorithm using variant allele frequency, chromosomal structural abnormalities, clinical and laboratory data and longitudinal data. This distinction is particularly important when evaluating minimal residual disease and in the presence of other neoplastic processes.
VEXAS Syndrome: Recently described VEXAS (vacuoles, E1 enzyme, X-linked, autoinflammatory, somatic) is caused by mutations in the UBA1 gene. This is an adults-onset fatal disease that may present as myelodysplastic syndrome, aplastic anemia or multiple myeloma, but is also characterized by fevers, low white cell count, vacuoles in bone marrow cells, dysplastic bone marrow, pulmonary inflammation, chondritis, and vasculitis. Detecting the presence of mutations in the UBA1 gene is the only way to confirm the diagnosis of this syndrome.
Acute Lymphoblastic Leukemia (ALL): This comprehensive assay is designed to confirm the diagnosis of Ph+ (BCR::ABL1-positive) ALL and Ph-like (BCR::ABL1-like) ALL and distinguish them from other types of ALL. It can be used for diagnosis as well as for monitoring. The incidence of Ph-like ALL is 20% to 25% of adult ALL and 15% of pediatric ALL. Diagnosis of Ph+-ALL and Ph-like ALL is very important because TKI therapy can be helpful in most of these patients. This assay can determine most of the mutations, translocations, and expression of genes (CRLF2) associated with Ph+ ALL and Ph-like ALL.
Diffuse Large B-cell Lymphoma (DLBCL) and Other Types of Lymphoma or Plasma Cell Neoplasms: This assay can provide very valuable information for the management and monitoring of patients with DLBCL. It can distinguish between activated B-cell-like (ABC) and germinal center B-cell-like (GCB) and can help in the diagnosis of double hit lymphoma. The assay is also useful for other types of lymphoma, including follicular lymphoma and T-cell neoplasms, as well as multiple myeloma.
IgHV Mutation Status: Very important for prognosis and therapy selection in patients with chronic lymphocytic leukemia (CLL).
T-cell and B-cell Clonality Detection: The detection of T- and B-cell clonality is important because it can help diagnose and monitor certain types of malignancies. When a malignant transformation occurs in a T- or B-cell, the cells can undergo uncontrolled clonal expansion, resulting in the accumulation of a large number of identical cells with the same T- or B-cell receptor.
Epstein-Barr Virus (EBV): Important for diagnosis and classification of lymphoproliferative neoplasms.
Torque Teno Virus (TTV): This virus was first discovered in a patient with non-A-E hepatitis and is now regarded as a part of the human virome. In general, TTV does not cause pathology in immunocompetent individuals. This virus is considered as a marker of the degree of immune competence in patients with immunological impairment and inflammatory disorders. High TTV load is associated with increased risk of infection. In patients with organ transplant, low TTV load is associated with an increased risk of rejection.
Other Features of Hematology Profile Plus:
- HLA class I genotyping
-Enrichment for CD138-positive plasma cells is performed on bone marrow specimens with the clinical indication of multiple myeloma
-A preliminary report will be issued for any patient positive for FLT3-ITD by fragment analysis or with a clinical indication of acute leukemia
GTC Accepts Skin Biopsies: Cutaneous lymphoid lesions are frequently encountered in skin biopsies. Differentiating between various benign and malignant entities is critical, as is a precise classification of cutaneous lymphoid infiltrate remains a vexing problem for clinicians and pathologists.
Turn Around Time: 7-10 Days
GTC uses AI in every step of our analysis and it makes a difference in helping make a new discovery daily that improves patient care.
Once the data is offloaded from the sequencer, our AI:
- Assists with mutation analysis, identifying non-mutations and artifacts
- Compares various data sets to explore disease biology
- Provides support for clinical decision-making and classification of the disease
- It helps with matching patients to therapeutics and presents clinical trial options
- Aggregates data for report generation and simplifies the results so they are easily understood
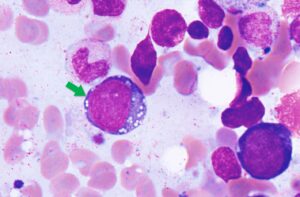
Case Study: VEXAS Syndrome
Background
VEXAS syndrome is a disease with inflammatory and hematologic (blood) manifestations. The syndrome is caused by mutations in the UBA1 gene affecting the Met41 residue of the protein and resulting in decreased cellular ubiquitylation activity and hyperinflammation. This is an adults-onset fatal disease that may present as myelodysplastic syndrome, aplastic anemia or multiple myeloma, but characterized by fevers, low white cell count, vacuoles in bone marrow cells, dysplastic bone marrow, pulmonary inflammation, chondritis, and vasculitis. Detecting UAB1 gene mutations in the is the only way for confirming the diagnosis of this disease.
Clinical History
68-year-old male
With low-grade myelodysplastic syndrome and inflammatory manifestations including arthritis, chondritis or other autoimmune disorders
Molecular Profiling Findings
Mutations in UBA1, DNMT3A, and TET2 genes
Discussion
The presence of DNMT3A, and TET2 suggest early low-grade myelodysplastic syndrome (MDS). And the detection of UBA1 mutation along with the patient’s symptoms confirmed the diagnosis of VEXAS. This patient was undiagnosed for long period of time and became transfusion dependent. By using any of our hematology DNA, DNA+RNA or cfDNA NGS panels; we are able to accurately diagnose VEXAS syndrome. This patient and most of the patients who are currently diagnosed with VEXAS have had numerous tests and tried multiple treatments. VEXAS should be considered in patients with systemic autoinflammatory disorders as well as patients with clinical presentation of myelodysplastic syndrome.
Download PDF (VEXAS case study brochure)
References
1. David B. Beck, M.D., et al. Somatic Mutations in UBA1 and Severe Adult-Onset Autoinflammatory Disease N Engl J Med 2020; 383:2628-2638, DOI: 10.1056/NEJMoa2026834
2. James A. Poulter, et al. Novel somatic mutations in UBA1 as a cause of VEXAS syndrome. Blood (2021) 137 (26): 3676–3681.
3. Marcela A. Ferrada, et al. Somatic Mutations in UBA1 Define a Distinct Subset of Relapsing Polychondritis Patients With VEXAS, Arithritis & Rheumatology, doi.org/10.1002/art.41743.
- Bone marrow: 2 mL. EDTA tube is preferred.
- Peripheral blood: 5-10 mL. EDTA tube is preferred.
- Fresh Tissue
- FFPE: 1 H&E slide and 8-10 unstained slides, 5-7 microns of BM clot or tissue fixed with 10% NBF fixative. Alternatively, the FFPE block of the BM clot can be sent for sectioning in our Lab.
- GTC Accepts skin biopsies
Specimen Preparation and Shipping Guidelines
Use the Hematology Transport Kit or Solid Tumor Transport Kit (depending on the sample type)
- Complete Requisition, making sure all sections are completed in their entirety including client information, patient Information, specimen Information and test Selection. Missing information may delay reporting of test results.
- Diagnosis/patient history is extremely important in rendering the correct interpretation of results and should also be filled out as completely as possible. A copy of a Path report should be included.
- Ensure the specimen is labeled with patient name and number. A minimum of two patient identifiers is required for each specimen
For FFPE samples:
- If sending FFPE Block-Insert up to 6 blocks into plastic block tray provided. Insert block tray into foam insert in the transport box.
- If sending slides-Insert slides into plastic slide holders provided. Insert the slide holders into the foam insert in the transport box. The container will hold up to 6 maximum slide holders.
- Place folded test requisition(s) and/or manifest(s) if ordered electronically into the transport kit.
- Close box and tuck tabs into place. No tape necessary.
For blood samples:
- Ship using a cold pack. The cold pack should not directly contact the blood tube. Ship as soon as sample collected with overnight delivery.
Request Kits
Fill out the form below to request kits. Please refer to the Specimen Requirements page for more details.
*GTC will need to set you up in our system if this is your first order.
How to complete the Genomic Testing Cooperative requisition form.
Download our
Test Requisition
Keep in mind that we do not accept blood samples directly from individuals. Talk with your M.D. to fill out the form for you.
Do You Want to Download the Sample Report?
Do You Want to Download the Sample Report?