A Pan-Tumor Assay
for Hematologic Malignancies
Pan-Tumor Assay for Hematologic Malignancies
GTC’s Liquid Trace Hematology is a highly sensitive, pan-cancer test that evaluates circulating cell-free (cf) RNA and DNA and provides highly informative data that can be used for diagnosis, evaluation of the host immune response, and identification of biomarkers useful for predicting response to various therapies.
GTC’s Liquid Trace can significantly reduce the need for bone marrow biopsies for hematology patients. Furthermore, the test can detect chromosomal abnormalities, translocations, and gene amplifications.
Liquid Trace can detect all types of hematologic cancers including:
- Multiple myeloma (MM)
- Lymphoma
- Acute lymphoblastic leukemia (ALL)
- Acute myeloid leukemia (AML)
- Myelodysplastic syndrome (MDS)
- Chronic myelomonocytic leukemia (CMML)
- Myeloproliferative neoplasm (MPN)
- Monitor therapeutic response
- VEXAS syndrome
- EBV-related neoplasms
- Hypereosinophilia
Many conventional liquid biopsy tests are dependent solely on cfDNA analysis, which presents multiple challenges. These include variations in DNA shedding between tumors as well as low sensitivity (especially in early-stage cancer), difficulty in detecting fusion genes (i.e., chromosomal translocations leading to the expression of chimeric mRNA from two genes), and inability to detect the numerous biological processes that modify RNA expression levels, such as alternative splicing, stability, and allele-specific methylation. The latter limitation is critically important as recent studies have shown that RNA testing provides another level of biological information regarding the tumor and its microenvironment.
The Benefits of cfRNA
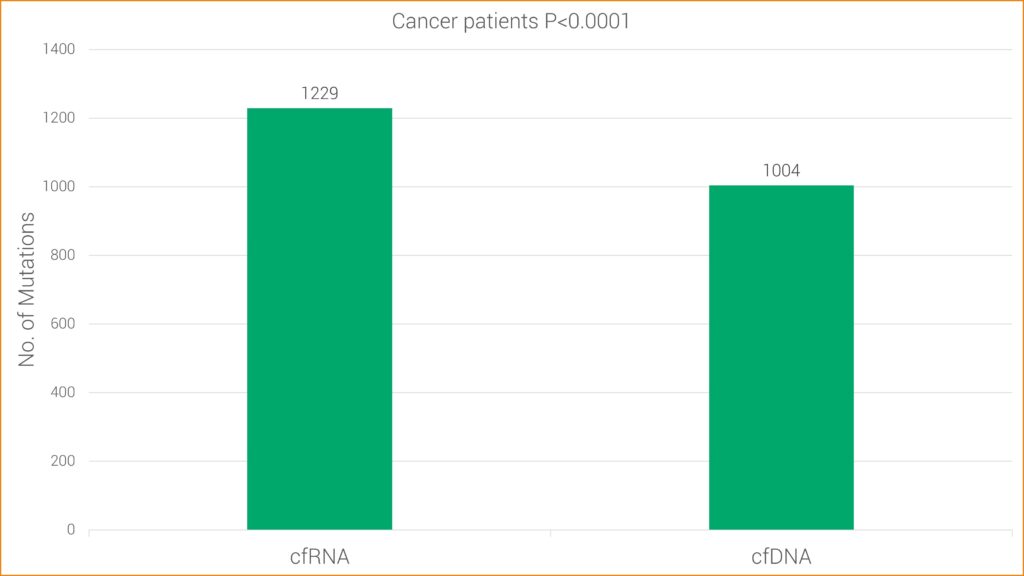
Studies have found RNA sequencing to be more sensitive for some types of mutations, likely because cancer cells typically contain one copy of mutated DNA but numerous copies of RNA. This research is consistent with GTC’s findings that cfRNA has increased sensitivity over cfDNA alone. More specifically, cfRNA allows GTC’s Liquid Trace to detect more mutations and fusions in hematologic and solid tumor samples, which may be undetected with conventional cfDNA.
T-cell and B-cell clonality detection: The detection of T- and B-cell clonality is important because it can help diagnose and monitor lymphoid and plasma cell malignancies. When malignant transformation occurs in T- or B-cells, the cells can undergo uncontrolled clonal expansion, resulting in the accumulation of many cells with the identical T- or B-cell receptor.
HLA class I genotyping: May be useful when identifying candidates for immunotherapy.
Torque teno virus (TTV): TTV was first discovered in a patient with non-A-E hepatitis and is now regarded as a part of the human virome. In general, it does not cause pathology in immunocompetent individuals. TTV is considered as a marker of the level of immune competence in patients with immunological impairment and inflammatory disorders. High TTV load is associated with increased risk of infection. In patients with organ transplant, low TTV load is associated with an increased risk of rejection.
Sensitivity is 0.1 to 0.01 for non-hot spot, 0.01 to 0.001 for hotspot and <0.001 for tumor informed or prior Hx.
For DNA, QNS is rare (<0.1%), but it is higher for RNA (Good DNA results but poor RNA results. Of course, if we receive 3 ml of plasma (6 ml blood), the sample is QNS for performing RNA testing.
VAF (Variant Allele Frequency) value: This value is used to monitor the disease in liquid bx. The high the VAF means higher tumor load. Patients showing reduction in VAF after treatment means they are doing better.
GTC uses AI in every step of our analysis, leading to daily discoveries that can help improve patient care.
Once the data are offloaded from the sequencer, our AI:
- Assists with mutation analysis, identifying non-mutations and artifacts
- Compares various data sets to explore disease biology
- Provides support for clinical decision making and classification of the disease
- Helps with matching patients to therapeutics and presents clinical trial options
- Aggregates data for report generation and simplifies the results so they are easily understood
Case Study: Multiple Myeloma
Background
Multiple myeloma is a malignancy originating in plasma cells. It is characterized by bone marrow infiltration, excessive production of monoclonal proteins, bone destruction, plasma cell aggregates, renal involvement, and associated anemia and immunodeficiency.
Multiple myeloma is heterogeneous in its genetic abnormalities and shows molecular evolution during disease progression and treatment. In addition, there is molecular diversity between subclones in the same or different bone marrow (BM) sites, between BM and the extramedullary sites, and between individuals. These features may explain why, despite improving and longer overall median survival over the past few years, some patients survive for decades whereas others succumb to the disease rapidly. As a result of these characteristics, optimal management of this malignancy requires testing modalities capable of capturing a comprehensive profile of the mutational and chromosomal landscape of the disease, enabling new risk stratification systems and individual therapy options.
Liquid biopsy has emerged as a promising non-invasive diagnostic tool used frequently in solid tumors but rarely in multiple myeloma. Bone marrow biopsy supplemented by flow cytometry, immunohistochemistry, cytogenetics, and fluorescence in situ hybridization (FISH), has traditionally been the gold standard for diagnosis and follow-up of multiple myeloma patients. However, liquid biopsy is emerging as a one-stop source, capable of not only supplementing, but increasingly replacing and surpassing many facets of traditional bone marrow biopsy. It offers several advantages over traditional bone marrow biopsy for evaluation of multiple myeloma, including but not limited to ease of sampling; real-time monitoring; identification of potential molecular targets and drug resistance; enhanced detection of chromosomal abnormalities, translocations, gene amplifications; and assessment of B-cell and T-cell clonality.
Liquid biopsy also has the potential to be used to assess measurable residual disease because it provides information on B-cell clonality as well as mutations.
Clinical History
• 61-year-old male with hypercalcemia, anemia, and new lytic bone lesions involving the left iliac bone and the right 9th rib.
• The patient had a free kappa light chain level of 1195 mg/L and a free lambda light chain level of 3.2 mg/L, with a kappa to lambda ratio of 373. Immunofixation showed an IgG monoclonal protein with kappa light chain specificity.
Molecular Profiling Findings
• Mutations in KMT2C (2 mutations), PTPN11 (2 mutations), WHSC1, PRKAR1A, TP53 (low level), CCNE1, TSHR, POLD1, EGFR, and DNMT3A genes.
• No detectable autosomal chromosomal structural gain or loss by copy-number variation (CNV) analysis.
• Increased plasma cell mRNA markers CD138 and BCMA.
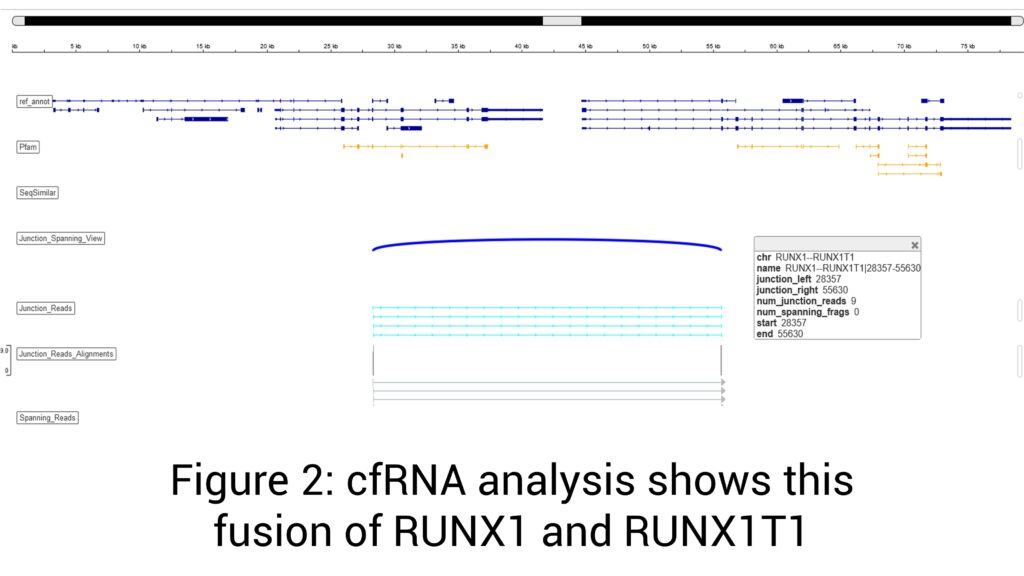
• Marked increase in CCND1 mRNA, reflecting promoter hijacking characteristic of t(11;14) CCND1-IgH.
• No evidence of high-risk chromosomal abnormalities consisting of t(4;14)(FGFR3/NSD2), t(14;16)(MAF), t(14;20)(MAFB), 1q21 + , del(1p), and del(17p) by RNA fusion or CNV analysis.
• No evidence of mutations in KRAS, NRAS, ATM, ATR, MYC, or DIS3.
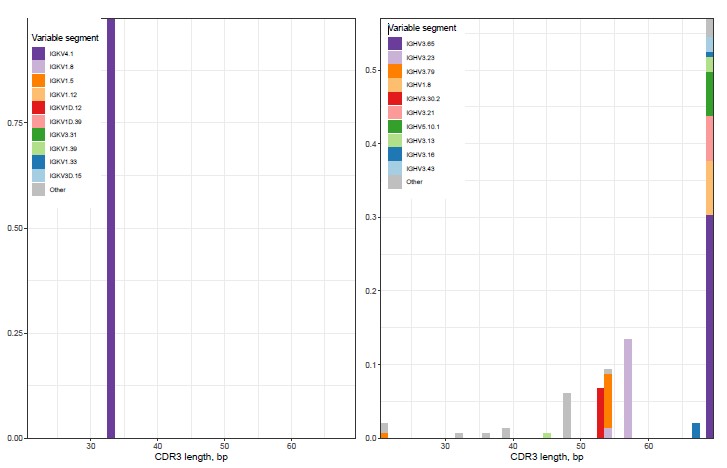
• Positive B-cell clonality detection (IgHV3.65/IgKV8.61); see figure 1.
Discussion
The mutational profile, elevated mRNA marker levels for plasma cells, and a positive B-cell clonality test are diagnostic of multiple myeloma.
The detection of t (11;14) CCND1-IgH by fusion mRNA is considered an intermediate risk category in multiple myeloma and confers a worse outcome compared to standard-risk myeloma. In addition, t(11;14) myeloma cases have been shown to be particularly sensitive to BCL-2 inhibitors, making BCL-2 a potential target in this subtype of myeloma.
This case also showed absence of poor prognostic chromosomal abnormalities consisting of t(4;14), t(14;16), t(14;20), 1q21 +, del(1p), and del(17p), as well as high-risk mutations consisting of KRAS, NRAS, ATM, ATR, MYC, and DIS3.
The aggregate findings in this case provide comprehensive genomic profiling that is superior to single-site tissue biopsy for diagnosis, prognostic profiling, treatment, and longitudinal management, given it minimally invasive nature and better representation of up-to-date tumor genome abnormalities and tumor genomic diversity.
Download Case Study Brochure (PDF)
References
1. Davies FE, Walker BA. What is genomic high-risk myeloma? Hemato. 2022; 3(2):287-297. https://doi.org/10.3390/hemato3020021
2. Ferreira, B., Caetano, J., Barahona, F. et al. Liquid biopsies for multiple myeloma in a time of precision medicine. J Mol Med 98, 513–525 (2020). https://doi.org/10.1007/s00109-020-01897-9
3. Liu Y, Guo J, Yi Y, Gao X, Wen L, Duan W, Wen Z, Liu Y, Guan Y, Xia X, Ma L, Fu R, Liu L, Huang X, Ge Q, Lu J. Circulating tumor DNA: less Invasive, more representative method to unveil the genomic landscape of newly diagnosed multiple myeloma than bone marrow aspirates. Cancers (Basel). 2022 Oct 7;14(19):4914. doi: 10.3390/cancers14194914. PMID: 36230837; PMCID: PMC9563595.
4. Li S, Zhang E, Cai Z. Liquid biopsy by analysis of circulating myeloma cells and cell-free nucleic acids: a novel noninvasive approach of disease evaluation in multiple myeloma. Biomark Res. 2023 Mar 8;11(1):27. doi: 10.1186/s40364-023-00469-6. PMID: 36890597; PMCID: PMC9997021.
- Peripheral blood: 10 mL in an EDTA tube is required.
Important: RNA stability is 48-72 hours from blood draw. DNA stability is 7 days from blood draw. Samples received beyond 72 hours may include only DNA results.
- CSF: 7-10 mL is optimal (5 mL minimum).
Important: Ship as soon as possible (overnight). Do not use collection devices with anti-coagulants. Clear tubes.
Specimen Preparation and Shipping Guidelines
Use the Hematology Transport Kit
- Complete the requisition, making sure that all sections are completed in their entirety, including client information, patient Information, specimen information, and test selection. Missing information may delay reporting of test results.
- Diagnosis/patient history is extremely important in rendering the correct interpretation of results and should also be filled out as completely as possible. A copy of a pathology report should be included.
- Ensure the specimen is labeled with patient name and number. A minimum of two patient identifiers is required for each specimen.
For blood samples:
- Ship using a cold pack. The cold pack should not directly contact the blood tube. Ship as soon with overnight delivery as the sample is collected.
Important: RNA stability is 48-72 hours from blood draw. DNA stability is 7 days from blood draw. Samples received beyond 72 hours may include only DNA results.
Request Kits
Fill out the form below to request kits. Please refer to the Specimen Requirements page for more details.
*GTC will need to set you up in our system if this is your first order.

How to complete the Genomic Testing Cooperative requisition form.
Download our
Test Requisition
Keep in mind that we do not accept blood samples directly from individuals. Talk with your M.D. to fill out the form for you.
1. Rogers A, Joe Y, Manshouri T, Dey A, Jilani I, Giles F, Estey E, Freireich E, Keating M, Kantarjian H, Albitar M. Relative increase in leukemia-specific DNA in peripheral blood plasma from patients with acute myeloid leukemia and myelodysplasia. Blood. 2004;103:2799–801. 2. Nakamura S, Yokoyama K, Shimizu E, Yusa N, Kondoh K, Ogawa M, Takei T, Kobayashi A, Ito M, Isobe M, Konuma T, Kato S, Kasajima R, Wada Y, Nagamura-Inoue T, Yamaguchi R, Takahashi S, Imoto S, Miyano S, Tojo A. Prognostic impact of circulating tumor DNA status post-allogeneic hematopoietic stem cell transplantation in AML and MDS. Blood. 2019 Jun 20;133(25):2682-2695. doi: 10.1182/blood-2018-10-880690. 3. Paul Yeh, Michael Dickinson, Sarah Ftouni, Tane Hunter, Devbarna Sinha, Stephen Q. Wong, Rishu Agarwal, Ravikiran Vedururu, Kenneth Doig, Chun Yew Fong, Piers Blombery, David Westerman, Mark A. Dawson and Sarah-Jane Dawson. Molecular disease monitoring using circulating tumor DNA in myelodysplastic syndromes. Blood. 2017;129(12):1685-1690. 4. Albitar A, Ma W, DeDios I, Estella J, Ahn I, Farooqui M, Wiestner A, Albitar M. Using high-sensitivity sequencing for the detection of mutations in BTK and PLCγ2 genes in cellular and cell-free DNA and correlation with progression in patients treated with BTK inhibitors. Oncotarget. 2017 Mar 14;8(11):17936-17944. doi: 10.18632/oncotarget.15316. PMID: 28212557 5. Albitar F, Ma W, Diep K, De Dios I, Agersborg S, Thangavelu M, Brodie S, Albitar M. Deep Sequencing of Cell-Free Peripheral Blood DNA as a Reliable Method for Confirming the Diagnosis of Myelodysplastic Syndrome. Genet Test Mol Biomarkers. 2016 Jul;20(7):341-5. doi: 10.1089/gtmb.2015.0278. PMID: 27248906 6. Aljurf M, Abalkhail H, Alseraihy A, Mohamed SY, Ayas M, Alsharif F, Alzahrani H, Al-Jefri A, Aldawsari G, Al-Ahmari A, Belgaumi AF, Walter CU, El-Solh H, Rasheed W, Albitar M. Chimerism Analysis of Cell-Free DNA in Patients Treated with Hematopoietic Stem Cell Transplantation May Predict Early Relapse in Patients with Hematologic Malignancies. Biotechnol Res Int. 2016;2016:8589270. doi: 10.1155/2016/8589270. 7. Ma W, Kantarjian H, Zhang X, Jilani I, Sheikholeslami MR, Donahue AC, Ravandi F, Estey E, O’Brien S, Keating M, Giles FJ, Albitar M. Detection of nucleophosmin gene mutations in plasma from patients with acute myeloid leukemia: clinical significance and implications. Cancer Biomark. 2009;5(1):51-8. doi: 10.3233/CBM-2009-0583. PMID: 19242062 8. Yeh CH, Tseng R, Albitar M. Plasma-based detection of clonality in lymphoid malignancies. Eur J Haematol. 2009 Jun;82(6):450-3. doi: 10.1111/j.1600-0609.2009.01231.x. PMID: 19187275. 9. Ma W, Kantarjian H, Zhang X, Sun W, Buller AM, Jilani I, Schwartz JG, Giles F, Albitar M. Higher detection rate of JAK2 mutation using plasma. Blood. 2008 Apr 1;111(7):3906-7. doi: 10.1182/blood-2008-02-139188. PMID: 18362222. 10. Giles FJ, Albitar M. Plasma-based testing as a new paradigm for clinical testing in hematologic diseases. Expert Rev Mol Diagn. 2007 Sep;7(5):615-23. Review. PMID: 17892367. 11. Ma W, Tseng R, Gorre M, Jilani I, Keating M, Kantarjian H, Cortes J, O’Brien S, Giles F, Albitar M. Plasma RNA as an alternative to cells for monitoring molecular response in patients with chronic myeloid leukemia. Haematologica. 2007 Feb;92(2):170-5. PMID: 17296565. 12. Ma W, Kantarjian H, Jilani I, Gorre M, Bhalla K, Ottmann O, Giles F, Albitar M. Heterogeneity in detecting Abl kinase mutations and better sensitivity using circulating plasma RNA. Leukemia. 2006 Nov;20(11):1989-91. Epub 2006 Aug 24. PMID: 16932346. 13. Ma W, Jilani I, Gorre M, Keating M, Chan H, Tseng R, Kantarjian H, O’Brien S, Giles FJ, Albitar M. Plasma as a source of mRNA for determining IgV(H) mutation status in patients with chronic lymphocytic leukaemia. Br J Haematol. 2006 Jun;133(6):690-2. 14. Jilani I, Estey E, Manshuri T, Caligiuri M, Keating M, Giles F, Thomas D, Kantarjian H, Albitar M. Better detection of FLT3 internal tandem duplication using peripheral blood plasma DNA. Leukemia. 2003 Jan;17(1):114-9. 15. Kurtz DM, Green MR, Bratman SV, Scherer F, Liu CL, Kunder CA, Takahashi K, Glover C, Keane C, Kihira S, Visser B, Callahan J, Kong KA, Faham M, Corbelli KS, Miklos D, Advani RH, Levy R, Hicks RJ, Hertzberg M, Ohgami RS, Gandhi MK, Diehn M, Alizadeh AA.Noninvasive monitoring of diffuse large B-cell lymphoma by immunoglobulin high-throughput sequencing. Blood. 2015 Jun 11;125(24):3679-87.
Do You Want to Download the Sample Report?
Do You Want to Download the Sample Report?