Accuracy of predicting IgHV mutation status in chronic lymphocytic leukemia using RNA expression profiling and machine learning
The mutation status of the variable region of the immunoglobulin heavy chain (IgHV) represents one of the most widely established prognostic markers of chronic lymphocytic leukemia (CLL) (1,2). The somatic hypermutation status of the clonotypic IgHV, in particular, has been shown to underpin the risk stratification process and clinical decision-making for patients with CLL (3). The IgHV genes can be either mutated or unmutated in patients with CLL, with the latter having inferior outcomes with standard therapies (4).
This comprehension of CLL and IgHV mutation status has several clinical applications, including deducing the appropriate course of treatment for patients. An enhanced response to chemoimmunotherapy in CLL patients with mutated IgHV has been demonstrated with about 60% of patients with no evidence of disease with a plateau at 15 years (5). On the other hand, patients with unmutated IgHV have an overall inferior response to chemoimmunotherapy and a shorter time to next therapy as well as a lower overall
survival (4,6,7). Jain et al. [2018] found that higher variation levels in IgHV mutations were increasingly and significantly associated with better progression-free survival and overall survival in CLL patients treated with FCR (fludarabine, cyclophosphamide and rituximab) (8). These results were replicated in the CLL8 trial, reported on by Fischer et al. [2016], which reported a significant increase in long-term remissions and overall survival in patients with mutated IgHV CLL after receiving FCR (9).
Most of the current testing for IgHV is performed using Sanger sequencing of PCR-amplified clonal or rearranged IgH variable complementarity determining regions 3 (CDR3). Sangar sequencing involves two distinct steps to determine IgHV mutation status. Firstly, clonality is detected. Second, the gene is then sequenced using Sanger sequencing and compared to predetermined germline genes obtained through immunoglobulin databases (10-12).
Despite the success and widespread acceptance of Sanger sequencing for IgHV mutation status detection, this approach of testing is not without its limitations. There are a large number of techniques available to detect IgHV mutation status; hence, discrepancies between institutions are rife.
Moreover, when using PCR, there is a risk that an alternative transcript will be amplified, giving inaccurate results. A similar disadvantage arises regarding certain primers omitting subclones (13). It has also been reported that using framework-region primers does not a full-length transcript to be deduced, leading to inaccuracies when calculating the percentage similarity to the homologous germline V region sequence (14). Although the availability of the immunoglobulin databases is an initial advantage when determining IgHV mutation status; this variable also poses a limitation to detection methods due to a large number of inconsistencies between the data provided. Variations may also be present in the software programs adopted to calculate the overall percentage of nucleotide mutations (13). Furthermore, it is well established that more than one clone in the CLL cell population can be seen in almost 10% of cases (15). This makes it very difficult to obtain accurate evaluation of the mutation status using Sanger sequencing.
Using NGS in sequencing can overcome most of these problems, especially when long sequence is used and covered leader region along with the other framework regions (16). NGS methodology also allows the detection of various subclones and families involved in the neoplastic process. However, NGS introduces a different set of problems, specifically determining the overall mutation status when IgHV families mutated and others that are not mutated present in the same sample. Furthermore, the IgHV mutation status sequencing does not provide any information on the presence of mutations in oncogenic genes that are relevant for evaluating the aggressiveness of the neoplastic clone.
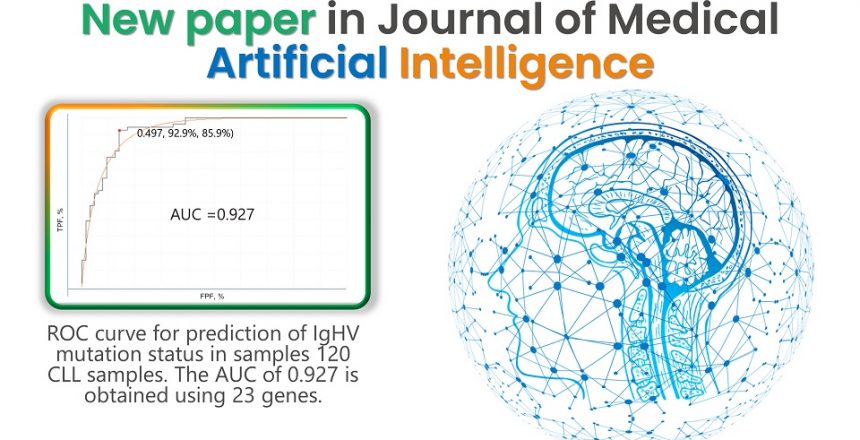
Here we describe the use of RNA expression profiling generated from routine targeted RNA sequencing by NGS along with machine learning for the prediction of the IgHV mutation status in patients with CLL. We present the following article in accordance with the STARD reporting checklist (available at https://jmai.amegroups.com/article/view/10.21037/jmai-22-28/rc).
To read the whole article download the paper.