Using high-sensitivity sequencing for the detection of mutations in BTK and PLCγ2 genes in cellular and cell-free DNA and correlation with progression in patients treated with BTK inhibitors.
Patients with chronic lymphocytic leukemia (CLL) that develop resistance to Bruton tyrosine kinase (BTK) inhibitors are typically positive for mutations in BTK or phospholipase c gamma 2 (PLCγ2). We developed a high sensitivity (HS) assay utilizing wild-type blocking polymerase chain reaction achieved via bridged and locked nucleic acids.
We used this high sensitivity assay in combination with Sanger sequencing and next generation sequencing (NGS) and tested cellular DNA and cell-free DNA (cfDNA) from patients with CLL treated with the BTK inhibitor, ibrutinib. We also tested ibrutinib-naïve patients with CLL. HS testing achieved 100x greater sensitivity than Sanger. HS Sanger sequencing was capable of detecting < 1 mutant allele in background of 1000 wild-type alleles (1:1000). Similar sensitivity was achieved with HS NGS. No BTK or PLCγ2 mutations were detected in any of the 44 ibrutinib-naïve CLL patients. We demonstrate that without the HS testing 56% of positive samples would have been missed for BTK and 85% of PLCγ2 would have been missed.
With the use of HS, we were able to detect multiple mutant clones in the same sample in 37.5% of patients; most would have been missed without HS testing. We also demonstrate that with HS sequencing, plasma cfDNA is more reliable than cellular DNA in detecting mutations. Our studies indicate that wild-type blocking and HS sequencing is necessary for proper and early detection of BTK or PLCγ2 mutations in monitoring patients treated with BTK inhibitors.
Furthermore, cfDNA from plasma is very reliable sample-type for testing.
[rule type=”basic”]
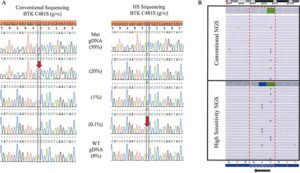
Reference Image A:
Increase in sensitivity with the addition of BNA/LNA oligonucleotides to sequencing
(A) Sanger Sequencing. Genomic DNA positive for a BTK C481S (g>c) mutation is serially diluted with WT DNA to determine the limit of detection for the conventional and high-sensitivity (HS) Sanger based assays. Limit of detection is approximately 20% mutant allele in a background of WT by the conventional assay compared with 0.1% in the HS assay. (B) Next Generation Sequencing. The lower panel shows the high sensitivity testing of a sample which clearly demonstrates three different DNA strands with three different mutations: c.1442G>C (C481S), c.1441T>C (C481R) and c.1441T>A (C481S). The upper panel shows the same sample tested in conventional NGS without the high sensitivity approach. The three mutations were detectable but at a lower frequency.
[rule type=”basic”]
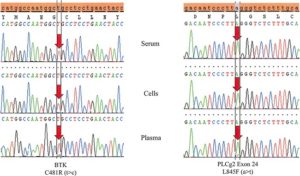
Reference Image B:
Testing DNA derived from peripheral blood plasma is more sensitive than serum and even cells
Temporally matched samples from the same patient (Patient # 4) reveals both a BTK C481R (t>c) and a PLCγ2 L845F (a>t) mutation in plasma. The BTK mutation is not detected in cells and both mutations are absent in serum.