The Liquid Biopsy Solid Tumor test uses next generation sequencing (NGS) performed on cell-free DNA (cfDNA) to identify molecular abnormalities in 277 genes implicated in solid tumors.
The Liquid Biopsy Solid Tumor is recommended for monitoring patients after establishing the molecular abnormalities using a baseline tissue sample. It can be used for detecting minimal residual disease (MRD) or the development of specific resistant mutations related to therapy (e.g. EGFR T790M mutation). We will analyze a tissue diagnostic sample, determine what genes are mutated in that baseline sample, then test the cfDNA for the specific mutations detected in the baseline sample. We will perform liquid biopsy testing for an initial diagnosis if tissue is unavailable.
Baseline tissue sample analysis is essential for proper molecular profiling of cancers. Tissue samples allow for the analysis of the RNA in addition to the DNA. RNA provides incredibly important information on expression pattern, overexpression, fusion genes, alternative splicing and mutations. We recommend tissue biopsy samples not only to establish baseline for monitoring, but for thorough evaluation including possible translocations that may involve ALK, ROS1, RET, NTRK, FGFR, etc. RNA testing allows for evaluation of the microenvironment (immune response, PD-L1, PD-L2, PD-1, CD8 cell, etc.). RNA analysis can also determine the MET exon 14 skipping and various alternative splicing and amplification of HER2, MET and EGFR. RNA information would be missing if the initial evaluation is performed by liquid biopsy alone.
Turn Around Time: 5-7 days
Peripheral blood: 5-10 mL. EDTA tube is preferred.
- Complete requisition, making sure all sections are completed in their entirety including client information, patient Information, specimen Information and test Selection. Missing information may delay reporting of test results.
- Diagnosis/patient History is extremely important in rendering the correct interpretation of results and should also be filled out as completely as possible.
- A copy of a path report should be included.
- Ensure the specimen is labeled with patient name and number. A minimum of two patient identifiers is required for each specimen.
Specimen Preparation and Shipping Guidelines
Use the Hematology Transport Kit
- Complete Requisition, making sure all sections are completed in their entirety including client information, patient Information, specimen Information and test Selection. Missing information may delay reporting of test results.
- Diagnosis/patient history is extremely important in rendering the correct interpretation of results and should also be filled out as completely as possible. A copy of a Path report should be included.
- Ensure the specimen is labeled with patient name and number. A minimum of two patient identifiers is required for each specimen.
For blood samples:
- Ship using a cold pack. The cold pack should not directly contact the blood tube. Ship as soon as sample collected with overnight delivery.
Genes validated and tested for Mutations in DNA testing for Solid Tumor
| 1-57 | 58-112 | 113-168 | 169-224 | 225-277 |
|---|---|---|---|---|
| ABL1 | CEBPA | FUBP1 | MLH1 | RB1 |
| ACVR1B | CHEK1 | GALNT12 | MPL | RET |
| AKT1 | CHEK2 | GATA1 | MRE11A | RHEB |
| AKT2 | CIC | GATA2 | MSH2 | RHOA |
| AKT3 | CREBBP | GATA3 | MSH6 | RIT1 |
| ALK | CRLF2 | GEN1 | MTOR | RNF43 |
| AMER1 | CSF1R | GNA11 | MUTYH | ROS1 |
| APC | CSF3R | GNAQ | MYC | RUNX1 |
| AR | CTCF | GNAS | MYCL | SDHB |
| ARAF | CTNNA1 | GREM1 | MYCN | SETBP1 |
| ARID1A | CTNNB1 | GRIN2A | MYD88 | SETD2 |
| ARID1B | CUX1 | H3F3A | NEF2 | SF3B1 |
| ARID2 | CXCR4 | HGF | NF1 | SMAD2 |
| ASXL1 | CYLD | HIST1H3B | NF2 | SMAD4 |
| ATM | DAXX | HNF1A | NFE2L2 | SMARCA4 |
| ATR | DDR2 | HOXB13 | NFKBIA | SMARCB1 |
| ATRX | DICER1 | HRAS | NKX2-1 | SMC1A |
| AURKA | DNM2 | HSP90AA1 | NOTCH1 | SMC3 |
| AURKB | DNMT3A | ID3 | NOTCH2 | SMO |
| AURKC | DOT1L | IDH1 | NOTCH3 | SOCS1 |
| AXIN1 | EED | IDH2 | NPM1 | SOX2 |
| AXIN2 | EGFR | IGF1R | NRAS | SOX9 |
| B2M | EGLN1 | IKZF1 | NSD1 | SPOP |
| BAP1 | EP300 | IKZF3 | NTRK1 | SRC |
| BCL2 | EPAS1 | IL7R | NTRK2 | SRSF2 |
| BCL2L1 | EPHA3 | INHBA | NTRK3 | STAG2 |
| BCL6 | EPHA5 | IRF4 | PAK3 | STAT3 |
| BCOR | ERBB2 | JAK1 | PALB2 | STK11 |
| BCORL1 | ERBB3 | JAK2 | PAX5 | SUFU |
| BCR | ERBB4 | JAK3 | PBRM1 | SUZ12 |
| BIRC3 | ERG | KAT6A | PDGFRA | TAL1 |
| BLM | ESR1 | KDM5C | PDGFRB | TCF3 |
| BRAF | ETV6 | KDM6A | PHF6 | TERT |
| BRCA1 | EXO1 | KDR | PIK3CA | TET2 |
| BRCA2 | EZH2 | KEAP1 | PIK3R1 | TGFBR2 |
| BRIP1 | FAM175A | KIT | PIK3R2 | TNFAIP3 |
| BTK | FAM46C | KMT2A | PIM1 | TNFRSF14 |
| CALR | FANCA | KMT2B | PLCG1 | TP53 |
| CARD11 | FANCC | KMT2C | PMS1 | TRAF3 |
| CBL | FANCD2 | KMT2D | PMS2 | TSC1 |
| CBLB | FANCE | KRAS | POLD1 | TSC2 |
| CBLC | FANCF | LRP1B | POLE | TSHR |
| CCND1 | FANCG | MAP2K1 | PPM1D | U2AF1 |
| CCND3 | FAS | MAP2K2 | PPP2R1A | U2AF2 |
| CCNE1 | FBXW7 | MAP2K4 | PRDM1 | UBA1 |
| CD274 | FGF4 | MAP3K1 | PRKAR1A | VHL |
| CD79A | FGF6 | MAP3K14 | PRKDC | WHSC1 |
| CD79B | FGFR1 | MAPK1 | PRSS1 | WT1 |
| CDC73 | FGFR2 | MCL1 | PTCH1 | XPO1 |
| CDH1 | FGFR3 | MDM2 | PTEN | XRCC2 |
| CDK12 | FGFR4 | MDM4 | PTPN11 | XRCC3 |
| CDK4 | FH | MED12 | RAC1 | ZNF217 |
| CDK6 | FLCN | MEF2B | RAD21 | ZRSR2 |
| CDKN2A | FLT3 | MEN1 | RAD50 | |
| CDKN2B | FLT4 | MET | RAD51 | |
| CDKN2C | FOXL2 | MITF | RAF1 |
Request Kits
Fill out the form below to request kits. Please refer to the Specimen Requirements page for more details.
*GTC will need to set you up in our system if this is your first order.
GTC uses AI in every step of our analysis and it makes a difference in helping make a new discovery daily that improve patient care.
Once the data is offloaded from the sequencer, our AI:
- Assists with mutation analysis, identifying non-mutations and artifacts
- Compares various data sets to explore disease biology
- Provides support for clinical decision making and classification of the disease
- It helps with matching patients to therapeutics and presents clinical trial options
- Aggregates data for report generation and simplifies the results so they are easily understood
Case Study: Liquid Biopsy cfDNA
Utility of this technology in clinical oncology
Background
Cell-free DNA (cfDNA) has become a reframing technology in clinical oncology. The ability to evaluate the presence, level, and constitution of tumor DNA from a routine, noninvasive blood draw has opened the door to a broad array of multiple clinical applications. Here we highlight the value of cfDNA molecular profiling along with the incorporation of the patient’s history for a complete landscape of the molecular features and interpretation of this information by experts.
Clinical History
- 78-year-old female
- With anemia,
- neutropenia, and
- history of carcinosarcoma of the uterus
Molecular Profiling Findings
- Germline mutation in CHEK2 gene, heterozygous
- Mutations in: DNMT3A (2 mutations), JAK2 (V617F), ARID1A, PTEN (2 mutations), KMT2D (2 mutations), KDM5C, AKT1, SETD2, TET2, PIK3R1, FGFR1, BCR, PIK3CA, TSC2, KMT2C, and DDR2 genes
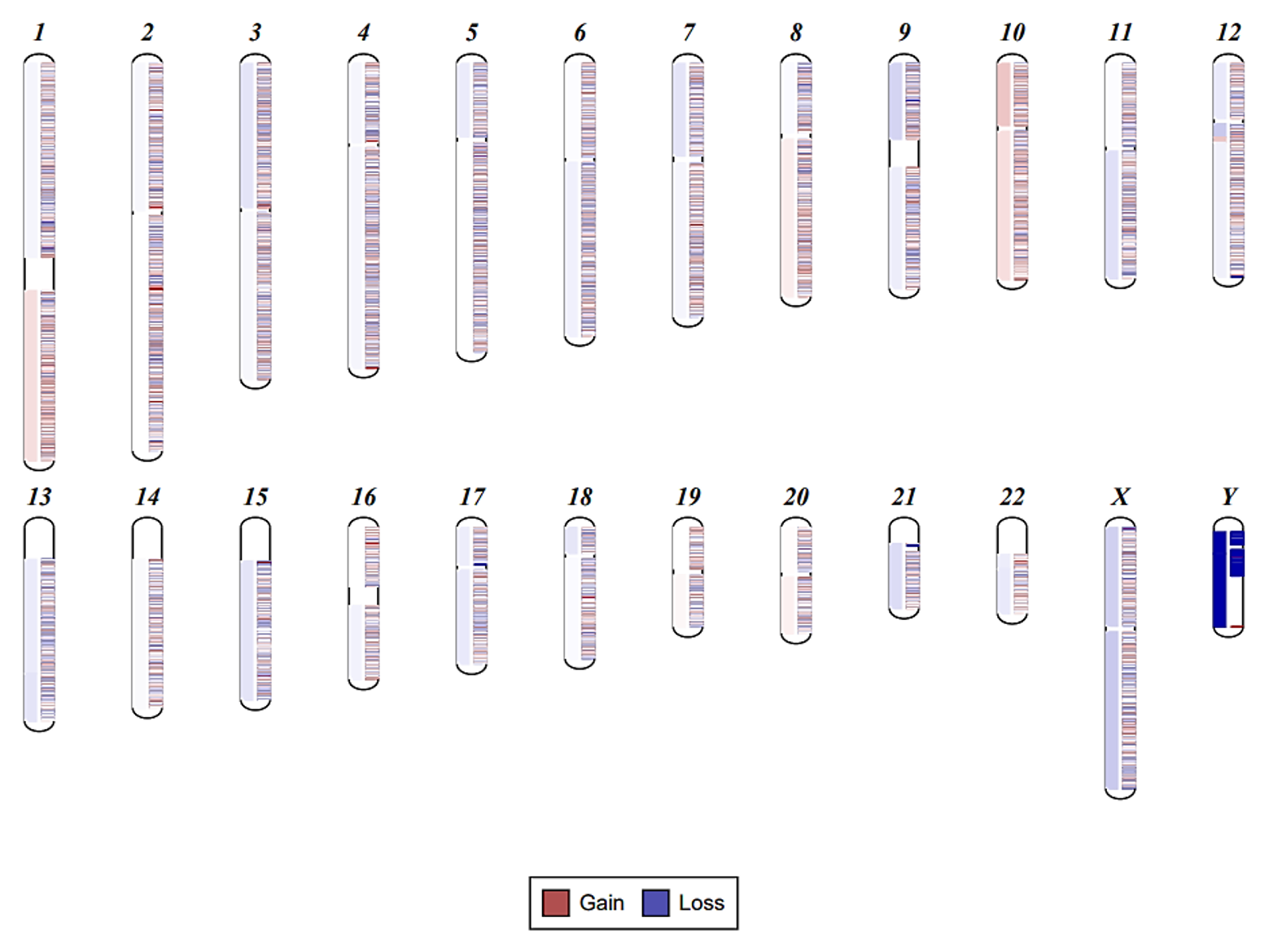
Chromosomal structural analysis showed1q+ and +10 (Figure1)
Discussion
The presence of CHEK2, DNMT3A (2 mutations), JAK2 (V617F), and TET2 is consistent with chronic myeloproliferative neoplasm (MPN). The detection of the other mutations along with the chromosomal changes highlights the presence of circulating solid tumor. Taken together, the findings in this case support the co-presence of chronic myeloproliferative neoplasm along with circulating tumor DNA with high tumor burden. While MPNs typically present with elevated white and red blood cell counts in this case the patient presented with anemia and neutropenia. The patient’s manifestations of MPN were masked by the fact that the patient was on chemotherapy and the routine CBC monitoring wouldn’t be able to detect it in a timely manner. By monitoring the patient’s cfDNA we were able to diagnose the patient and detect her tumor burden promptly and offer her the optimal supportive and prophylactic therapy.
cfDNA testing is also used in the clinic for mutational profiling. Integrating real-time cfDNA monitoring into clinical care provides invaluable opportunities to precisely track the course of a patient’s response to therapy, and it could allow not just the early detection of imminent cancer progression but also the identification of the molecular mechanisms driving resistance or disease progression.
Therapeutic Recommendations
- PIK3CA mutation suggests response to PI3K/mTOR inhibitors.
- PTEN, ARID1A and CHEK2 mutations suggest response to PARP inhibitors.
- AKT1 mutation suggests possible response to AKT inhibitors
- FGFR1 mutation suggests response to FGFR1 inhibitor
*See full list of therapeutics in the drug information section of the report
Download Case Study Brochure (PDF)
References
1. Albitar M, Ip A, Goy A, Estella J, De Dios I, Ma W, Pecora A, Koprivnikar J and McCloskey J. Reliability of Cell-Free DNA (cfDNA) Next
Generation Sequencing in Predicting Chromosomal Structural Abnormalities and Cytogenetic-Risk Stratification of Patients with Myeloid
Neoplasms, AMERICAN SOCIETY OF HEMATOLOGY. Oral and Poster Abstracts 2021
2. Greenfield G, McMullin M & Mills K. Molecular pathogenesis of the myeloproliferative neoplasms. J Hematol Oncol 14, 103 (2021).
https://doi.org/10.1186/s13045-021-01116-z
3. Bronkhorst AJ, Ungerer V, Holdenrieder S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol
Detect Quantif. 2019;17:100087. Published 2019 Mar 18. doi:10.1016/j.bdq.2019.100087
How to complete the Genomic Testing Cooperative requisition form.
Download our
Test Requisition
Keep in mind that we do not accept blood samples directly from individuals. Talk with your M.D. to fill out the form for you.
Do You Want to Download the Sample Report?
Hematology
Profile Plus- TAT
7-10 Days - Indications
Classification and diagnosis of lymphoma, multiple myeloma, acute lymphoblastic leukemia, acute myeloid leukemia
Includes IgVH
- - Sample Type
Bone marrow,
Peripheral blood,
Fresh tissue - Sample Requirements
Bone marrow: 2ml.
Peripheral blood: 5 ml.
EDTA tube preferred
FFPE: 1 H&E slide and 6-10 unstained slides, 5-7 microns of tissue fixed with 10% NBF fixative - Results Reported
DNA+RNA - Diagnostic
- Therapeutic
- Prognostic
- Heterogeneity
- Clinical Trial Matching